Abstract
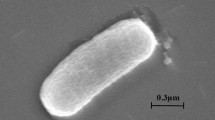
A Gram-negative, aerobic, non-motile, rod-shaped, floc-forming, and non-spore-forming bacterium, designated as NLF-7-7T, was isolated from the biofilm of a sample collected from a livestock wastewater treatment plant in Nonsan, Republic of Korea. Strain NLF-7-7T, forms a visible floc and grows in the flocculated state. Cells of strain NLF-7-7T grew optimally at pH 6.5 and 30 °C and in the presence of 0.5% (w/v) NaCl. Phylogenetic analysis based on 16S rRNA gene sequences revealed that strain NLF-7-7T belonged to the family Comamonadaceae, and was most closely related to Comamonas badia DSM 17552T (95.8% similarity) and Comamonas nitrativorans 23310T (94.0% similarity). The phylogenetic and phenotypic data indicate strain NLF-7-7T is clearly distinguished from the Comamonas lineage. The major cellular fatty acids were C10:0 3OH, C16:0, and summed feature 3 (C16:1 ω6c/C16:1 ω7c). The respiratory quinone was Q-8. The polar lipids were composed of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and an unidentified aminolipid. The DNA G+C content of strain NLF-7-7 was 68.0 mol%. Based on the phenotypic, chemotaxonomic, and phylogenetic properties, strain NLF-7-7T represents a novel species of the genus Comamonas, for which the name Comamonas flocculans sp. nov. is proposed. The type strain is C. flocculans NLF-7-7T (=KCTC 62943T). The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of Comamonas flocculans NLF-7-7T is MN527436. The whole-genome shotgun BioProject Number is PRJNA555370 with the Accession Number CP042344.

Similar content being viewed by others
Abbreviations
- AL:
-
Aminolipid
- DPG:
-
Diphosphatidylglycerol
- HPLC:
-
High-performance liquid chromatography
- KCTC:
-
Korean Collection for Type Cultures
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- TEM:
-
Transmission electron microscopy
- TLC:
-
Thin-layer chromatography
References
Davis GH, Park RW (1962) A taxonomic study of certain bacteria currently classified as Vibrio species. J Gen Microbiol 27:101–119
Euzéby JP (2018) LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Bacteriol 68:1825–1829
Wauters G, De Baere T, Willems A, Falsen E, Vaneechoutte M (2003) Description of Comamonas aquatica comb. nov. and Comamonas kerstersii sp. nov. for two subgroups of Comamonas terrigena and emended description of Comamonas terrigena. Int J Syst Evol Microbiol 53:859–862
Gumaelius L, Magnusson G, Pettersson B, Dalhammar G (2001) Comamonas denitrificans sp. nov., an efficient denitrifying bacterium isolated from activated sludge. Int J Syst Evol Microbiol 51:999–1006
Tago Y, Yokota A (2004) Comamonas badia sp. nov., a floc-forming bacterium isolated from activated sludge. J Gen Appl Microbiol 50:243–248
Young CC, Chou JH, Arun AB, Yen WS, Sheu SY, Shen FT, Lai WA, Rekha PD, Chen WM (2008) Comamonas composti sp. nov., isolated from food waste compost. Int J Syst Evol Microbiol 58:251–256
Kim KH, Ten LN, Liu QM, Im WT, Lee ST (2008) Comamonas granuli sp. nov., isolated from granules used in a wastewater treatment plant. J Microbiol 46:390–395
Zhang J, Wang Y, Zhou S, Wu C, He J, Li F (2013) Comamonas guangdongensis sp. nov., isolated from subterranean forest sediment, and emended description of the genus Comamonas. Int J Syst Evol Microbiol 63:809–814
Sun LN, Zhang J, Chen Q, He J, Li QF, Li SP (2013) Comamonas jiangduensis sp. nov., a biosurfactant-producing bacterium isolated from agricultural soil. Int J Syst Evol Microbiol 63:2168–2173
Bidault A, Clauss F, Helaine D, Balavoine C (1997) Floc agglomeration and structuration by a specific talc mineral composition. Water Sci Technol 36:57–68
Eikelboom DH, van Buijsen HJJ (1983) Microscopic sludge investigation manual, 2nd edn. TNO Institute of Environmental Health, Delft, pp 1–100
Nielsen JL, Juretschko S, Wagner M, Nielsen PH (2002) Abundance and phylogenetic affiliation of iron reducers in activated sludge as assessed by fluorescence in situ hybridization and microautoradiography. Appl Environ Microbiol 68:4629–4636
Wagner M, Loy A, Nogueira R, Purkhold U, Lee N, Daims H (2002) Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek 81:665–680
Friedman BA, Dugan PR (1968) Identification of Zoogloea species and the relationship to zoogloeal matrix and floc formation. J Bacteriol 95:1903–1909
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Minnikin D, O’Donnell A, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Tindall B (1989) Fully saturated menaquinones in the archaebacterium Pyrobaculum islandicum. FEMS Microbiol Lett 60:251–253
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI, Inc., Newark
Kim DH, Han KI, Kwon HJ, Kim MG, Kim YG, Choi DH, Lee KC, Suh MK, Kim HS, Lee JS, Kim JG (2019) Complete genome sequence of Comamonas sp. NLF-7-7 isolated from biofilter of wastewater treatment plant. Korean J Microbiol 55:309–312
Acknowledgements
We thank Professor Bernhard Schink for his advice in naming the novel strain.
Funding
This research was supported by the Project for Cooperative R&D between Industry, Academy, and Research Institute funded by the Korea Ministry of SMEs and Startups in 2018 (Grant Number S2597396); and the Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, DH., Han, KI., Kwon, HJ. et al. Comamonas flocculans sp. nov., a Floc-Forming Bacterium Isolated from Livestock Wastewater. Curr Microbiol 77, 1902–1908 (2020). https://doi.org/10.1007/s00284-020-01940-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01940-5