Abstract
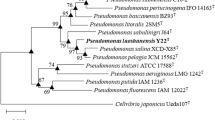
A Gram-negative, strictly aerobic, motile, rod-shaped bacterium with monopolar flagellum, designated as F51T, was isolated from the skin ulcer of farmed Murray cod sampled from Zhejiang Province, China. Strain F51T grew at 4–37 °C (optimal temperature, 28 °C), pH 5.0–8.5 (optimal pH, 7.5) and NaCl concentration of 0–6.0% (w/v) (optimal concentration, 2.0%). Phylogenetic analysis based on average nucleotide identity (76.2–78.4%) and in silico DNA–DNA hybridization (22.3–23.2%) values revealed that strain F51T forms a distinct lineage in the clade of genus Pseudomonas with less than 98.9% 16S rRNA gene sequence similarity to type strains of the genus and represents a novel species related most closely to Pseudomonas floridensis LMG 30013T. Three housekeeping genes (rpoB, rpoD and gyrB) of strain F51T were analysed to further confirm that the isolate is distinctly delineated from related Pseudomonas species. Chemotaxonomic analysis indicated that the sole respiratory quinone of strain F51T is Q-9; its predominant cellular fatty acids are C16:0, summed feature 3 (iso-C15:0 2-OH and/or C16:1ω7c), summed feature 8 (C18:1ω7c and/or C18:1ω6c) and C10:0 3-OH; and its major polar lipids consist of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, two unidentified glycolipids, three unidentified phospholipids and an unidentified aminophosphoglycolipid. This composition is typical of the chemotaxonomic attributes of Pseudomonas. Based on its phenotypic, chemotaxonomic and phylogenetic features, strain F51T is considered to represent a novel species for which the name Pseudomonas ovata sp. nov. is proposed. The type strain is F51T (= KCTC 62133T = MCCC 1K03458T).

Similar content being viewed by others
References
Migula W (1894) Über ein neues System der Bakterien. Arb Bakteriol Inst Karlsruhe 1:235–238
Sudan SK, Pal D, Bisht B, Kumar N, Chaudhry V, Patil P, Sahni G, Mayilraj S, Krishnamurthi S (2017) Pseudomonas fluvialis sp. nov., a novel member of the genus Pseudomonas isolated from the river Ganges, India. Int J Syst Evolut Microbiol 68:402–408
Peix A, Ramírez-Bahena MH, Velázquez E (2018) The current status on the taxonomy of Pseudomonas revisited: an update. Infect Genet Evolut 57:106–116
Liu YC, Young LS, Lin SY, Hameed A, Hsu YH, Lai WA, Shen FT, Young CC (2013) Pseudomonas guguanensis sp. nov., a gammaproteobacterium isolated from a hot spring. Int J Syst Evolut Microbiol 63(Pt 12):4591–4598
Frasson D, Opoku M, Picozzi T, Torossi T, Balada S, Thm S, Hilber U (2017) Pseudomonas wadenswilerensis sp. nov. and Pseudomonas reidholzensis sp. nov., two novel species within the Pseudomonas putida group isolated from forest soil. Int J Syst Evolut Microbiol 67(8):2853–2861
Lin SY, Hameed A, Liu YC, Hsu YH, Lai WA, Chen WM, Shen FT, Young CC (2013) Pseudomonas sagittaria sp. nov., a siderophore-producing bacterium isolated from oil-contaminated soil. Int J Syst Evolut Microbiol 63(7):2410–2417
Toro M, Ramírez-Bahena MH, Cuesta MJ, Velázquez E, Peix A (2013) Pseudomonas guariconensis sp. nov., isolated from rhizospheric soil. Int J Syst Evolut Microbiol 63(Pt 12):4413–4420
Pascual J, Lucena T, Ruvira MA, Giordano A, Gambacorta A, Garay E, Arahal DR, Pujalte MJ, Macián MC (2012) Pseudomonas litoralis sp. nov., isolated from Mediterranean seawater. Int J Syst Evolut Microbiol 62(2):438–444
Kim KH, Roh SW, Chang HW, Nam YD, Yoon JH, Che OJ, Oh HM, Bae JW (2009) Pseudomonas sabulinigri sp. nov., isolated from black beach sand. Int J Syst Evolut Microbiol 59(1):38–41
Liu R, Liu H, Feng H, Wang X, Zhang CX, Zhang KY, Lai R (2008) Pseudomonas duriflava sp. nov., isolated from a desert soil. Int J Syst Evolut Microbiol 58(6):1404–1408
de Bentzmann S, Plésiat P (2011) The Pseudomonas aeruginosa opportunistic pathogen and human infections. Environ Microbiol 13(7):1655–1665
Hirano SS, Upper CD (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64(3):624–653
Zdorovenko EL, Cimmino A, Marchi G, Shashkov AS, Fiori M, Knirel YA, Evidente A (2017) Studies on the O-specific polysaccharide of the lipopolysaccharide from the Pseudomonas mediterranea strain C5P1radi, a bacterium pathogenic of tomato and chrysanthemum. Carbohydr Res 448:48–51
Chen WP, Kuo TT (1993) A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res 21(9):2260
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evolut Microbiol 67(5):1613–1617
Liu Y, Rao QH, Tu JF, Zhang JN, Huang MM, Hu B, Lin Q, Luo TY (2018) Acinetobacter piscicola sp. nov., isolated from diseased farmed Murray cod (Maccullochella peelii peelii). Int J Syst Evolut Microbiol 68:905–910
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evolut 33(7):1870–1874
Kamjam M, Sivalingam P, Deng ZX, Hong K (2017) Deep sea Actinomycetes and their secondary metabolites. Front Microbiol 8:760
Tayeb LA, Ageron E, Grimont F, Grimont PA (2005) Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res Microbiol 156(5):763–773
Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S (2000) Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146(10):2385–2394
Mulet M, Lalucat J, Garcíavaldés E (2010) DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol 12(6):1513–1530
Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdés E (2012) Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst Appl Microbiol 35(7):455–464
Bennasar A, Mulet M, Lalucat J, García-Valdés E (2010) PseudoMLSA: a database for multigenic sequence analysis of Pseudomonas species. BMC Microbiol 10(1):1–6
Gomila M, Peña A, Mulet M, Lalucat J, Garcíavaldés E (2015) Phylogenomics and systematics in Pseudomonas. Front Microbiol 6(214):214
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14(1):60
Lee I, Kim YO, Park SC, Chun J (2015) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evolut Microbiol 66(2):1100–1103
Chaudhary DK, Kim J (2017) Chryseobacterium nepalense sp. nov., isolated from oil-contaminated soil. Int J Syst Evolut Microbiol 67(3):646–652
Chen C, Su Y, Tao T, Fu G, Zhang C, Sun C, Zhang X, Wu M (2016) Maripseudobacter aurantiacus gen. nov., sp. nov., a novel genus of the family Flavobacteriaceae isolated from sedimentation basin. Int J Syst Evolut Microbiol 67(4):778–783
Collins MD (1985) Isoprenoid quinone analyses in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Komagata K, Suzuki KI (1987) Lipid and cell-wall analysis in bacterial systematics. Method Microbiol 19:161–207
Ramírezbahena MH, Cuesta MJ, Floresfélix JD, Mulas R, Rivas R, Castropinto J, Brañas J, Mulas D, Gonzálezandrés F, Velázquez E (2014) Pseudomonas helmanticensis sp. nov., isolated from forest soil. Int J Syst Evolut Microbiol 64(7):2338–2345
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25(7):1043–1055
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106(45):19126–19131
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Can Entomol 37(4):443–537
Konstantinidis KT, Tiedje JM (2005) Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187(18):6258–6264
Acknowledgements
This work was financially supported by the Natural Science Foundation of Fujian Province of China (No. 2017J01051), Aquaculture Technology Service Team (No. kjFW19), NDRC Investment Project of Fujian (No. [2016] 482) and the Agricultural Product Quality Safety Innovation Team Project of FAAS (No. STIT2017-1-12).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest.
Ethical Approval
All the experiments were performed in compliance with the Chinese legislation on the use and care of laboratory animals, and were approved by the Ethical Committee of experiments Animal Care at laboratory animal management office of Zhejiang province (Approval No: SYXK 2013-0191).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA, gyrB, rpoB and rpoD gene sequences of strain F51T are MH305533, MH684587, MH684586 and MH684588, and the DPD number is TA00862. Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession QFFU00000000. The version described in this paper is version QFFU01000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rao, Q., Liu, Y., Chen, C. et al. Pseudomonas ovata sp. nov., Isolated from the Skin of the Tail of Farmed Murray cod (Maccullochella peelii peelii) with a Profound Ulceration. Curr Microbiol 76, 1168–1174 (2019). https://doi.org/10.1007/s00284-019-01729-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01729-1