Abstract
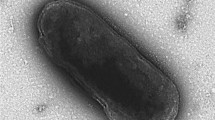
A novel Gram-stain-negative bacterium, designated strain BH-SD17T, was isolated from a marine sediment sample in the Bohai Gulf, Yellow Sea, China. Cells of strain BH-SD17T are aerobic, yellow-colored, non-flagellated rods. Growth occurs between 15 and 40 °C (optimum, 30 °C), at pH 6.0–8.5 (optimum, 7.5) and with 1.0–8.0% (w/v) NaCl (optimum, 3.0%). Strain BH-SD17T contains phosphatidylethanolamine and two unidentified lipids as the major polar lipids. The predominant fatty acids are iso-C15:0 (28.5%), iso-C15:1 G (24.4%), and iso-C17:0 3-OH (12.3%). The major respiratory quinone is MK-6. Strain BH-SD17T shows moderate 16S rRNA gene sequence similarity to existing identified strains, is most closely related to the genera Lutimonas (92.1–92.4%), Lutibacter (91.6–92.3%), and Taeania (91.9%). Phylogenetic trees based on 16S rRNA gene sequences show that strain BH-SD17T forms a distinct lineage within the family Flavobacteriaceae. Based on the results of phenotypic, chemotaxonomic and phylogenetic analysis, strain BH-SD17T is considered to represent a novel genus and species in the family Flavobacteriaceae, for which the name Aureibaculum marinum is proposed. The type strain is BH-SD17T (=CCTCC AB 2017072T=KCTC 62204T).

Similar content being viewed by others
References
Bernardet JF (2011) Family I. Flavobacteriaceae Reichenbach 1992. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, 2nd edn. Springer, New York, pp 106–111
Bernardet JF, Nakagawa Y, Holmes B (2002) Subcommittee on the taxonomy of Flavobacterium and Cytophaga-like bacteria of the International Committee on Systematics of Prokaryotes. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Choi DH, Cho BC (2006) Lutibacter litoralis gen. nov., sp. nov., a marine bacterium of the family Flavobacteriaceae isolated from tidal flat sediment. Int J Syst Evol Microbiol 56:771–776
Dong XZ, Cai MY (eds) (2001) Chapter 14. Determination of biochemical characteristics. In: Manual for the systematic identification of general bacteria. Science Press, Beijing, pp 370–398
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, Birol I (2017) ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res 27:768–777
Ji X, Zhang C, Zhang X, Xu Z, Ding Y, Zhang Y, Song Q, Li B, Zhao H (2018) Pelagivirga sediminicola gen. nov., sp. nov. isolated from the Bohai Sea. Int J Syst Evol Microbiol 68:3494–3499
Jung YT, Kim JH, Kang SJ, Oh TK, Yoon JH (2012) Namhaeicola litoreusgen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from seawater. Int J Syst Evol Microbiol 62:2163–2168
Jung YT, Yoon SY, Lee JS, Yoon JH (2016) Taeania maliponensis gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from seawater. Int J Syst Evol Microbiol 66:3552–3557
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbial 62:716–721
Kim YO, Park S, Nam BH, Jung YT, Kim DG, Bae KS, Yoon JH (2014) Description of Lutimonas halocynthiae sp. nov., isolated from a golden sea squirt (Halocynthia aurantium), reclassification of Aestuariicola saemankumensis as Lutimonas saemankumensis comb. nov. and emended description of the genus Lutimonas. Int J Syst Evol Microbiol 64:1984–1990
Komagata K, Suzuki KI (1988) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Park S, Park JM, Won SM, Park DS, Yoon JH (2015) Lutibacter crassostreae sp. nov., isolated from oyster. Int J Syst Evol Microbiol 65:2689–2695
Reichenbach H, Balows A, Trüper HG, Dworkin M, Harder W (1992) The order Cytophagales. In: Schleifer KH (ed) The Prokaryotes, 2nd edn. Springer, New York, pp 3631–3675
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Yang SJ, Choo YJ, Cho JC (2007) Lutimonas vermicola gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from the marine polychaete Periserrula leucophryna. Int J Syst Evol Microbiol 57:1679–1684
Yoon JH, Kang SJ, Jung YT, Oh TK (2008) Aestuariicola saemankumensis gen. nov., sp. nov., a member of the family Flavobacteriaceae, isolated from tidal flat sediment. Int J Syst Evol Microbiol 58:2126–2131
Acknowledgements
We sincerely thank Prof. Aharon Oren in Hebrew University of Jerusalem for advice on nomenclature. This work was supported by the National Natural Science Foundation of China (81702054, 81672044, 81501718) and Natural Science Foundation of Shandong Province (ZR2017BC011).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
All authors declare no conflicts of interest.
Ethical Statement
This article does not contain any studies with human participants or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, H., Wu, Y., Zhang, C. et al. Aureibaculum marinum gen. nov., sp. nov., a Novel Bacterium of the Family Flavobacteriaceae Isolated from the Bohai Gulf. Curr Microbiol 76, 975–981 (2019). https://doi.org/10.1007/s00284-019-01691-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01691-y