Abstract
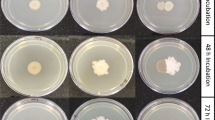
Successful colonization is the initial step for plant-bacteria interactions; therefore, the development of strategies to improve adherence to plant surfaces is critically important for environmental bacteria. Biofilm formation is thought to be one such strategy for bacteria to establish stable colonization on inert and living surfaces. Although biofilms play potential roles in enabling persistent bacterial colonization, little attention has been paid to biofilms formed by plant-associated bacteria. In this study, we characterized the biofilm-forming ability of 6 species of bacteria from the family Pseudomonadaceae: Pseudomonas protegens, Pseudomonas fluorescens, Pseudomonas putida, Pseudomonas stutzeri, Pseudomonas mendocina, and Pseudomonas syringae. These strains exhibit different degrees of biofilm formation depending on incubation time and nutrient availability. Distinct preferences for growth media were observed, as biofilms were formed by P. protegens with rich nutrients and by P. fluorescens and P. putida with poor nutrients. Likewise, P. stutzeri did not form biofilms with rich nutrients but did form biofilms under nutrient-poor conditions. These observations indicate that particular components in media may influence biofilm formation. P. putida, one of the strains with high biofilm-forming ability, showed the highest ability for initial attachment, which may be mediated by the hydrophobicity of its cell surface. P. mendocina also has high ability for initial attachment, and this strain produces cell surface-attached extracellular polysaccharides that promote cell aggregation. Thus, each strain possesses different properties that facilitate biofilm formation. Shedding light on bacterial strategies for colonization via biofilm formation would enable a better understanding of plant–bacteria interactions.





Similar content being viewed by others
References
Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R (2013) Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA 110(17):E1621–E1630. doi:10.1073/pnas.1218984110
Davies D (2003) Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2(2):114–122. doi:10.1038/nrd1008
Davies DG, Marques CN (2009) A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191(5):1393–1403. doi:10.1128/JB.01214-08
Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280(5361):295–298
Dewanti R, Wong AC (1995) Influence of culture conditions on biofilm formation by Escherichia coli O157:h7. Int J Food Microbiol 26(2):147–164
Dunne WM Jr (2002) Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 15(2):155–166
Egamberdiyeva D (2005) Characterization of Pseudomonas species isolated from the rhizosphere of plants grown in serozem soil, semi arid region of Uzbekistan. Sci World J 5:501–509. doi:10.1100/tsw.2005.64
Friedman L, Kolter R (2004) Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51(3):675–690
Goller C, Wang X, Itoh Y, Romeo T (2006) The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol 188(23):8022–8032. doi:10.1128/JB.01106-06
Huynh TT, McDougald D, Klebensberger J, Al Qarni B, Barraud N, Rice SA, Kjelleberg S, Schleheck D (2012) Glucose starvation-induced dispersal of Pseudomonas aeruginosa biofilms is cAMP and energy dependent. PLoS ONE 7(8):e42874. doi:10.1371/journal.pone.0042874
Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ (2004) Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol 186(14):4466–4475. doi:10.1128/JB.186.14.4466-4475.2004
Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48(6):1511–1524
Lakshmanan V, Kitto SL, Caplan JL, Hsueh YH, Kearns DB, Wu YS, Bais HP (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160(3):1642–1661. doi:10.1104/pp.112.200386
Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S (2007) A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol 65(6):1474–1484. doi:10.1111/j.1365-2958.2007.05879.x
Loper JE, Kobayashi DY, Paulsen IT (2007) The genomic sequence of Pseudomonas fluorescens Pf-5: insights into biological control. Phytopathology 97(2):233–238. doi:10.1094/PHYTO-97-2-0233
Monds RD, Newell PD, Gross RH, O’Toole GA (2007) Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63(3):656–679. doi:10.1111/j.1365-2958.2006.05539.x
O’Toole G, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi:10.1146/annurev.micro.54.1.49
Osman SF, Fett WF, Fishman ML (1986) Exopolysaccharides of the phytopathogen Pseudomonas syringae pv. glycinea. J Bacteriol 166(1):66–71
Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS 3rd, Thomashow LS, Loper JE (2005) Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23(7):873–878. doi:10.1038/nbt1110
Rich JJ, Hirano SS, Willis DK (1992) Pathovar-specific requirement for the Pseudomonas syringae lemA gene in disease lesion formation. Appl Environ Microbiol 58(5):1440–1446
Ross P, Mayer R, Weinhouse H, Amikam D, Huggirat Y, Benziman M, de Vroom E, Fidder A, de Paus P, Sliedregt LA et al (1990) The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J Biol Chem 265(31):18933–18943
Ryder C, Byrd M, Wozniak DJ (2007) Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10(6):644–648. doi:10.1016/j.mib.2007.09.010
Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, Gilbert P (2004) Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186(21):7312–7326. doi:10.1128/JB.186.21.7312-7326.2004
Silby MW, Cerdeno-Tarraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang XX, Moon CD, Gehrig SM, Godfrey SA, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10(5):R51. doi:10.1186/gb-2009-10-5-r51
Ueda A, Attila C, Whiteley M, Wood TK (2009) Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb Biotechnol 2(1):62–74. doi:10.1111/j.1751-7915.2008.00060.x
Ueda A, Wood TK (2008) Potassium and sodium transporters of Pseudomonas aeruginosa regulate virulence to barley. Appl Microbiol Biotechnol 79(5):843–858. doi:10.1007/s00253-008-1483-5
Ueda A, Wood TK (2009) Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5(6):e1000483. doi:10.1371/journal.ppat.1000483
Ueda A, Wood TK (2010) Tyrosine phosphatase TpbA of Pseudomonas aeruginosa controls extracellular DNA via cyclic diguanylic acid concentrations. Environ Microbiol Rep 2(3):449–455. doi:10.1111/j.1758-2229.2010.00171.x
Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS (2002) Extracellular DNA required for bacterial biofilm formation. Science 295(5559):1487. doi:10.1126/science.295.5559.1487
Yan Y, Yang J, Dou Y, Chen M, Ping S, Peng J, Lu W, Zhang W, Yao Z, Li H, Liu W, He S, Geng L, Zhang X, Yang F, Yu H, Zhan Y, Li D, Lin Z, Wang Y, Elmerich C, Lin M, Jin Q (2008) Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA 105(21):7564–7569. doi:10.1073/pnas.0801093105
Zhang XS, Garcia-Contreras R, Wood TK (2007) YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol 189(8):3051–3062. doi:10.1128/JB.01832-06
Acknowledgments
We are grateful to Professors George A. O’Toole (P. protegens Pf-5, P. fluorescens Pf0-1), Juan Ruis Ramos (P. putida KT2440), Steven Lindow (P. syringae B728a) for kindly providing the strains. We are also grateful to NBRC (P. mendocina NBRC 14162) and Pasteur Institute (P. stutzeri A1501) for distributing the strains. This research was supported in part by the grants from Takano Life Science Research Foundation and the Institute for Fermentation, Osaka.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ueda, A., Saneoka, H. Characterization of the Ability to Form Biofilms by Plant-Associated Pseudomonas Species. Curr Microbiol 70, 506–513 (2015). https://doi.org/10.1007/s00284-014-0749-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0749-7