Abstract
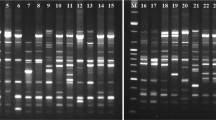
Pleurotus pulmonarius is one of the most widely cultivated and popular edible fungi in the genus Pleurotus. Three molecular markers were used to analyze the genetic diversity of 15 Chinese P. pulmonarius cultivars. In total, 21 random amplified polymorphic DNA (RAPD), 20 inter-simple sequence repeat (ISSR), and 20 sequence-related amplified polymorphism (SRAP) primers or primer pairs were selected for generating data based on their clear banding profiles produced. With the use of these RAPD, ISSR, and SRAP primers or primer pairs, a total of 361 RAPD, 283 ISSR, and 131 SRAP fragments were detected, of which 287 (79.5 %) RAPD, 211 (74.6 %) ISSR, and 98 (74.8 %) SRAP fragments were polymorphic. Unweighted Pair-Group Method with Arithmetic Mean (UPGMA) trees of these three methods were structured similarly, grouping the 15 tested strains into four clades. Subsequently, visual DNA fingerprinting and cluster analysis were performed to evaluate the resolving power of the combined RAPD, ISSR, and SRAP markers in the differentiation among these strains. The results of this study demonstrated that each method above could efficiently differentiate P. pulmonarius cultivars and could thus be considered an efficient tool for surveying genetic diversity of P. pulmonarius.




Similar content being viewed by others
References
Bao DP, Koike A, Yao FJ, Yamanaka K, Aimi T, Kitamoto Y (2007) Analyses of the genetic diversity of matsutake isolates collected from different ecological environments in Asia. J Wood Sci 53(4):344–350
Chang ST, Buswell JA (2008) Development of the world mushroom industry: applied mushroom biology and international mushroom organizations. Int J Med Mushrooms 10(3):195–208
Dai YC, Zhou LW, Yang ZL, Wen HA, Bau T, Li TH (2010) A revised checklist of edible fungi in China. Mycosystema 29(1):1–21
Depeiges A, Goubely C, Lenoir A, Cocherel S, Picard G, Raynal M, Grellet F, Delseny M (1995) Identification of the most represented repeated motifs in Arabidopsis thaliana microsatellite loci. Theor Appl Genet 91:160–168
Du P, Cui BK, Dai YC (2011) Genetic diversity of wild Auricularia polytricha in Yunnan Province of South-western China revealed by sequence-related amplified polymorphism (SRAP) analysis. J Med Plants Res 5(8):1374–1381
Fu LZ, Zhang HY, Wu XQ, Li HB, Wei HL, Wu QQ, Wang LA (2010) Evaluation of genetic diversity in Lentinula edodes strains using RAPD, ISSR and SRAP markers. World J Microb Biotech 26(4):709–716
Gepts P (1993) The use of molecular and biochemical markers in crop evolution studies. Evol Biol 27:51–94
Gregori A, Svagelj M, Pohleven J (2007) Cultivation techniques and medicinal properties of Pleurotus spp. Food Technol Biotech 45(3):238–249
Guan XJ, Xu L, Shao YC, Wang ZR, Chen FS, Luo XC (2008) Differentiation of commercial strains of Agaricus species in China with inter-simple sequence repeat marker. World J Microb Biotech 24(8):1617–1622
Huang NL, Lin ZB, Chen GL (2010) Medicinal and edible fungi. Shanghai science and technology literature press, Shanghai
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Loarce Y, Gallego R, Ferrer E (1996) A comparative analysis of genetic relationships between rye cultivars using RFLP and RAPD markers. Euphytica 88:107–115
Paterson AH, Tanksley SD, Sorrells ME (1991) DNA markers in plant improvement. Adv Agron 46:39–90
Ro HS, Kim SS, Ryu JS, Jeon CO, Lee TS, Lee HS (2007) Comparative studies on the diversity of the edible mushroom Pleurotus eryngii: ITS sequence analysis, RAPD fingerprinting, and physiological characteristics. Mycol Res 111:710–715
Rohlf FJ (2000) Exeter Software. NTSYS-pc: numerical taxonomy and multivariate analysis system. Setauket, New York
Sikdar B, Bhattacharya M, Mukherjee A, Banerjee A, Ghosh E, Ghosh B, Roy SC (2010) Genetic diversity in important members of Cucurbitaceae using isozyme. RAPD and ISSR markers. Biologia Plantarum 54(1):135–140
Sneath PHA, Sokal RR (1973) Taxonomic structure. In: Numerical taxonomy: the principles and practice of numerical classification. W.H. Freeman, San Francisco, CA, pp 188–308
Sun SJ, Gao W, Lin SQ, Zhu J, Xie BG, Lin ZB (2006) Analysis of genetic diversity in Ganoderma population with a novel molecular marker SRAP. Appl Microb Biotech 72(3):537–543
Tang LH, Xiao Y, Li L, Guo QA, Bian YB (2010) Analysis of genetic diversity among Chinese Auricularia auricula cultivars using combined ISSR and SRAP markers. Cur Microb 61(2):132–140
Urbanelli S, Della Rosa V, Punelli F, Porretta D, Reverberi M, Fabbri AA, Fanelli C (2007) DNA-fingerprinting (AFLP and RFLP) for genotypic identification in species of the Pleurotus eryngii complex. Appl Microbiol Biot 74(3):592–600
Wang SX, Yin YG, Liu Y, Xu F (2012) Evaluation of genetic diversity among chinese Pleurotus eryngii cultivars by combined RAPD/ISSR marker. Curr Microb 65(4):424–431
Yin YG, Liu Y, Wang SX, Zhao S, Xu F (2012) Examining genetic relationships of Chinese Pleurotus ostreatus cultivars by combined RAPD and SRAP Markers. Mycoscience 54(3):221–225
Yu MY, Ma B, Luo X, Zheng LY, Xu XY, Yang ZR (2008) Molecular diversity of Auricularia polytricha revealed by inter-simple sequence repeat and sequence-related amplified polymorphism markers. Cur Microb 56(3):240–245
Zervakis GI, Venturella G, Papadopoulou K (2001) Genetic polymorphism and taxonomic infrastructure of the Pleurotus eryngii species-complex as determined by RAPD analysis, iso-zyme profiles and ecomorphological characters. Microbiol (SGM) 147:3183–3194
Zhang M, Zhu L, Cui SW, Wang Q, Zhou T, Shen HS (2011) Fractionation, partial characterization and bioactivity of water-soluble polysaccharides and polysaccharide-protein complexes from Pleurotus geesteranus. Int J Biol Macromol 48(1):5–12
Zhang RY, Huang CY, Zheng SY, Zhang JX, Ng TB, Jiang RB, Zuo XM, Wang HX (2007) Strain-typing of Lentinula edodes in China with inter simple sequence repeat markers. Appl Microb Biotech 74(1):140–145
Acknowledgments
We thank the reviewers for the helpful comments and improvement of the text. This study was supported by grants from the Beijing Nova Program (Grant No. 2011053), the Beijing Academy of Agriculture and Forestry Science (Grant No. KJCX201101002), and Beijing Innovative Team of Edible Fungi, China Agriculture Research System (Grant No. PXM2012-036204-00153).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yin, Y., Liu, Y., Li, H. et al. Genetic Diversity of Pleurotus pulmonarius Revealed by RAPD, ISSR, and SRAP Fingerprinting. Curr Microbiol 68, 397–403 (2014). https://doi.org/10.1007/s00284-013-0489-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0489-0