Abstract
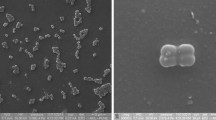
The taxonomic position of a Gram-positive, endo-spore forming bacterium isolated from a haematite ore sample was analyzed by a polyphasic approach. The strain designated as HIO-4T matched most of the phenotypic and chemical characteristics of the genus Cohnella and represents a novel species. The sequence of the almost complete 16S rRNA (1489 bases) was compared with those of previously studied Cohnella type strains and confirmed that the strain belongs to the genus Cohnella. Strain HIO-4T differs from all other species of Cohnella by at least 3.9% at the 16S rRNA level and the moderately related species are Cohnella phaseoli (96.1%) and Cohnella yongneupensis (96.1%), respectively. Predominant polar lipids are diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylethanolamine (PE); few unknown phospholipids, mannose containing lipid, aminophospholipid and aminophosphoglycolipids. The results of physiological and biochemical tests allowed the genotypic and phenotypic distinctiveness of strain HIO-4T with its phylogenetic relatives and suggest that the strain HIO-4T should be recognized as a novel species, for which the name Cohnella ferri sp. nov. is proposed. The type strain is HIO-4T (=MTCC 8365T = JCM 16139T)


Similar content being viewed by others
References
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Cai F, Wang Y, Qi H, Dai J, Yu B, An H, Rahman E, Fang C (2010) Cohnella luojiensis sp. nov., isolated from soil of a Euphrates poplar forest. Int J Syst Evol Microbiol 60:1605–1608
Cho EA, Lee JS, Lee KC, Jung HC, Pan JG, Pyun YR (2007) Cohnella laeviribosi sp. nov., isolated from a volcanic pond. Int J Syst Evol Microbiol 57:2902–2907
Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Cowan ST, Steel KJ (1965) Manual for the identification of medical bacteria. Cambridge University Press, London
De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB (2009) Cohnella Bergey’s manual of systematic bacteriology, 2nd edn. Springer, New York, pp 316–319
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
GarcıaFraile P, Velazquez E, Mateos PF, Molina EM, Rivas R (2008) Cohnella phaseoli sp. nov., isolated from root nodules of Phaseolus coccineus in Spain, and emended description of the genus Cohnella. Int J Syst Evol Microbiol 58:1855–1859
Kämpfer P, RossellóMora R, Falsen E, Busse HJ, Tindall BJ (2006) Cohnella thermotolerans gen. nov., sp. nov., and classification of ‘Paenibacillus hongkongensis’ as Cohnella hongkongensis sp. nov. Int J Syst Evol Microbiol 56:781–786
Khianngam S, Tanasupawat S, Akaracharanya A, Kim KK, Lee KC, Lee JS (2010) Cohnella thailandensis sp. nov., a xylanolytic bacterium from Thai soil. Int J Syst Evol Microbiol 60:2284–2287
Khianngam S, Tanasupawat S, Akaracharanya A, Kim KK, Lee KC, Lee JS (2010) Cohnella xylanilytica sp. nov. and Cohnella terrae sp. nov., xylanolytic bacteria from soil. Int J Syst Evol Microbiol 60:2913–2917
Kim JS, Weon HY, Kim YS, Anandham R, Jeon YA, Hong SB, Kwon SW (2009) Cohnella yongneupensis sp. nov. and Cohnella ginsengisoli sp. nov., isolated from two different soils. Int J Syst Evol Microbiol 60:526–530
Komagata K, Suzuki K (1987) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–206
Lanyi B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Luo X, Wang Z, Dai J, Zhang L, Fang C (2010) Cohnella damensis sp. nov., a motile xylanolytic bacteria isolated from a low altitude area in Tibet. J Microbiol Biotechnol 20(2):410–414
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the gunine plus cytosine content of DNA. Methods Enzymol 12B:195–206
Mayilraj S, Prasad GS, Suresh K, Saini HS, Shivaji S, Chakrabarti T (2005) Planococcus stackebrandtii sp. nov., isolated from a cold desert of the Himalayas, India. Int J Syst Evol Microbiol 55:91–94
Mayilraj S, Kroppenstedt RM, Suresh K, Saini HS (2006) Kocuria himachalensis sp. nov., a novel actinobacterium isolated from the Himalayas, India. Int J Syst Evol Microbiol 56:1971–1975
Minnikin DE, O′Donnell AG, Goodfellow M, Alderson G, Athaly M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Murray RG, Doetsch RN, Robinow CF (1994) Determinative and cytological light microscopy. In: Gerhard P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 21–41
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical note 101. MIDI Inc, Newark, DE
Shiratori H, Tagami Y, Beppu T, Ueda K (2009) Cohnella fontinalis sp. nov., a xylanolytic bacterium isolated from fresh water. Int J Syst Evol Microbiol 60:1344–1348
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhard P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Smith NR, Gordon RE, Clark FE (1952) Aerobic spore forming bacteria. U.S. Department of Agriculture Agricultural Monograph, no. 16
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wayne LG, Brenner DJ, Colwell RR, 9 other authors (1987) International Committee on Systematic Bacteriology. Report of the adhoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Acknowledgments
We thank Professor J P Euzéby, Ecole Nationale Veterinaire, France for his suggestion on Latin nomenclature for the novel species. I would like to thank Ms Anju Choudhary, Ms Deepika Sharma and Mr Malkit Singh for their excellent technical assistance and Ms Varpreet Kaur for getting the iron ore samples. AR is a recipient of DBT Postdoctoral fellowship. Financial assistance from CSIR and DBT, Government of India is duly acknowledged. This is IMTECH communication number 084/2010.
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank accession number for the 16S rRNA sequence of strain HIO-4T is EF 203083.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mayilraj, S., Ruckmani, A., Kaur, C. et al. Cohnella ferri sp. nov. A Novel Member of the Genus Cohnella Isolated from Haematite Ore. Curr Microbiol 62, 1704–1709 (2011). https://doi.org/10.1007/s00284-011-9917-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-011-9917-1