Abstract
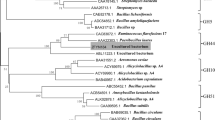
Metagenomics, a new research field developed over the past decade, aims to identify potential enzymes from nonculturable microbes. In this study, genes encoding three glycoside hydrolase family (GHF) 9 endoglucanases and one GHF 5 endoglucanase were cloned and identified from the metagenome of the compost soils. The shared identities between the predicted amino acid sequences of these genes and their closest homologues in the database were less than 70%. One GHF 9 endoglucanase, Umcel9B, was further characterized. The recombinant protein, Umcel9B, showed activity against carboxymethyl cellulose, indicating that Umcel9B is an endoactive enzyme. Enzymatic activity occurs optimally at a pH of 7.0 and a temperature of 25°C.


Similar content being viewed by others
References
Arai T, Kosugi A, Chan H et al (2006) Properties of cellulosomal family 9 cellulases from Clostridium cellulovorans. Appl Microbiol Biotechnol 71:654–660
Bhat MK (2000) Cellulases and related enzymes in biotechnology. Biotechnol Adv 18:355–383
Daniel R (2004) The soil metagenome—a rich resource for the discovery of novel natural products. Curr Opin Biotechnol 15:199–204
Feller G, Gerday C (2003) Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol 1:200–208
Feng Y, Duan CJ, Pang H et al (2007) Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl Microbiol Biotechnol 75:319–328
Ferrer M, Golyshina O, Beloqui A et al (2007) Mining enzymes from extreme environments. Curr Opin Microbiol 10:207–214
Hakamada Y, Endo K, Takizawa S et al (2002) Enzymatic properties, crystallization, and deduced amino acid sequence of an alkaline endoglucanase from Bacillus circulans. Biochim Biophys Acta 1570:174–180
Iyo AH, Forsberg CW (1999) A cold-active glucanase from the ruminal bacterium Fibrobacter succinogenes S85. Appl Environ Microbiol 65:995–998
Kim SJ, Lee CM, Han BR et al (2008) Characterization of a gene encoding cellulase from uncultured soil bacteria. FEMS Microbiol Lett 282:44–51
Lynd LR, Weimer PJ, van Zyl WH et al (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
McHardy AC, Rigoutsos I (2007) What’s in the mix: phylogenetic classification of metagenome sequence samples. Curr Opin Microbiol 10:499–503
Miettinen-Oinonen A, Suominen P (2002) Enhanced production of Trichoderma reesei endoglucanases and use of the new cellulase preparations in producing the stonewashed effect on denim fabric. Appl Environ Microbiol 68:3956–3964
Patrick W, Joey W, Alison R et al (2003) A rapid, cost-effective procedure for the extraction of microbial DNA from soil. World J Microb Biot 19:85–91
Rappe MS, Giovannoni SJ (2003) The uncultured microbial majority. Annu Rev Microbiol 57:369–394
Rondon MR, August PR, Bettermann AD et al (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Shimonaka A, Koga J, Baba Y et al (2006) Specific characteristics of family 45 endoglucanases from Mucorales in the use of textiles and laundry. Biosci Biotechnol Biochem 70:1013–1016
Teather RM, Wood PJ (1982) Use of Congo red-polysaccharide interactions in enumeration and characterisation of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol 43:777–780
Voget S, Leggewie C, Uesbeck A et al (2003) Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol 69:6235–6242
Voget S, Steele HL, Streit WR (2006) Characterization of a metagenome-derived halotolerant cellulase. J Biotechnol 126:26–36
Zeng R, Xiong P, Wen J (2006) Characterization and gene cloning of a cold-active cellulase from a deep-sea psychrotrophic bacterium Pseudoalteromonas sp. DY3. Extremophiles 10:79–82
Acknowledgment
This work was supported by Grants for New Century Excellent Talents in Universities of China (NCET-05-0752), the National Natural Science Foundation of China (30560003) and Hi-tech Research and Development Program of China (863 Program 2007AA021307).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pang, H., Zhang, P., Duan, CJ. et al. Identification of Cellulase Genes from the Metagenomes of Compost Soils and Functional Characterization of One Novel Endoglucanase. Curr Microbiol 58, 404–408 (2009). https://doi.org/10.1007/s00284-008-9346-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-008-9346-y