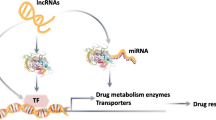
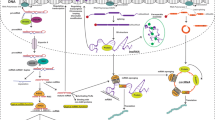
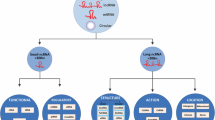
Abstract
Like almost all cancer types, timely diagnosis is needed for leukemias to be effectively cured. Drug efflux, attenuated drug uptake, altered drug metabolism, and epigenetic alterations are just several of the key mechanisms by which drug resistance develops. All of these mechanisms are orchestrated by up- and downregulators, in which non-coding RNAs (ncRNAs) do not encode specific proteins in most cases; albeit, some of them have been found to exhibit the potential for protein-coding. Notwithstanding, ncRNAs are chiefly known for their contribution to the regulation of physiological processes, as well as the pathological ones, such as cell proliferation, apoptosis, and immune responses. Specifically, in the case of leukemia chemo-resistance, ncRNAs have been recognized to be responsible for modulating the initiation and progression of drug resistance. Herein, we comprehensively reviewed the role of ncRNAs, specifically its effect on molecular mechanisms and signaling pathways, in the development of leukemia drug resistance.




Similar content being viewed by others
Data availability
Not applicable.
References
Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG (2013) Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13(10):714–726
Short NJ, Konopleva M, Kadia TM, Borthakur G, Ravandi F, DiNardo CD et al (2020) Advances in the treatment of acute myeloid leukemia: new drugs and new challenges advances in AML therapeutics. Cancer discovery 10(4):506–525
Crick FH (1958) On protein synthesis. Symp Soc Exp Biol 12:138–63
Kimura T (2020) Non-coding natural antisense RNA: mechanisms of action in the regulation of target gene expression and its clinical implications. Yakugaku Zasshi 140(5):687–700
Wong CM, Tsang FH, Ng IO (2018) Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 15(3):137–151
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE et al (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772):901–906
Shahrouki P, Larsson E (2012) The non-coding oncogene: a case of missing DNA evidence? Front Genet 3:170
Saw PE, Xu X, Chen J, Song EW (2021) Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci 64(1):22–50
Zhang P, Wu W, Chen Q, Chen M (2019) Non-coding RNAs and their integrated networks. J Integr Bioinform 16(3):20190027
Panni S, Lovering RC, Porras P, Orchard S (1863) Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech 2020(6):194417
Yan H, Bu P (2021) Non-coding RNA in cancer. Essays Biochem 65(4):625–639
Mirzaei HR, Sahebkar A, Mohammadi M, Yari R, Salehi H, Jafari MH, Namdar A, Khabazian E, Jaafari MR, Mirzaei H (2016) Circulating microRNAs in Hepatocellular Carcinoma: Potential Diagnostic and Prognostic Biomarkers. Current pharmaceutical design 22(34):5257–5269. https://doi.org/10.2174/1381612822666160303110838
De Vincentis A, Rahmani Z, Muley M, Vespasiani-Gentilucci U, Ruggiero S, Zamani P, Jamialahmadi T, Sahebkar A (2020) Long noncoding RNAs in nonalcoholic fatty liver disease and liver fibrosis: state-of-the-art and perspectives in diagnosis and treatment. Drug discovery today 25(7):1277–1286. https://doi.org/10.1016/j.drudis.2020.05.009
Gorabi AM, Ghanbari M, Sathyapalan T, Jamialahmadi T, Sahebkar A (2021) Implications of microRNAs in the Pathogenesis of Atherosclerosis and Prospects for Therapy. Current drug targets 22(15):1738–1749. https://doi.org/10.2174/1389450122666210120143450
Anastasiadou E, Jacob LS, Slack FJ (2018) Non-coding RNA networks in cancer. Nat Rev Cancer 18(1):5–18
E Nicolas F. Role of ncRNAs in development, diagnosis and treatment of human cancer. Recent Pat Anticancer Drug Discov 2017;12(2):128-135
Salehi M, Vafadar A, Khatami SH, Taheri-Anganeh M, Vakili O, Savardashtaki A et al Gastrointestinal cancer drug resistance: the role of exosomal miRNAs. Mol Biol Rep 49(3):2421–2432
Movahedpour A, Khatami SH, Khorsand M, Salehi M, Savardashtaki A, Mirmajidi SH et al (2021) Exosomal noncoding RNAs: key players in glioblastoma drug resistance. Mol Cell Biochem 476:4081–4092
Fawzy MS, Toraih EA, El-Wazir A, Hosny MM, Badran DI, El Kelish A (2019) Long intergenic non-coding RNA, regulator of reprogramming (LINC-ROR) over-expression predicts poor prognosis in renal cell carcinoma. Arch Med Sci 17(4):1016–1027. https://doi.org/10.5114/aoms.2019.85201
Han W, Niu L, Wang L, Liu J, Li H (2019) Downregulation of long non-coding RNA B-Raf proto-oncogene-activated non-coding RNA reverses cisplatin resistance in laryngeal squamous cell carcinoma. Archives of medical science: AMS 17(5):1164–1174. https://doi.org/10.5114/aoms.2019.91352
Han C, Yang Y, Guo L, Guan Q, Ruan S (2021Mar 18) The expression of long non-coding RNA HOTAIR in advanced hepatocellular carcinomaand its prognostic correlation with sunitinib therapy. Arch Med Sci 18(1):71–78. https://doi.org/10.5114/aoms/100480
Zhang S, Wang X, Wang D (2019) Long non-coding RNA LINC01296 promotes progression of oral squamous cell carcinoma through activating the MAPK/ERK signaling pathway via the miR-485-5p/PAK4 axis. Archives of medical science : AMS 18(3):786–799. https://doi.org/10.5114/aoms.2019.86805
Corrà F, Agnoletto C, Minotti L, Baldassari F, Volinia S (2018) The network of non-coding RNAs in cancer drug resistance. Front Oncol 8:327
Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X (2013) MicroRNA: function, detection, and bioanalysis. Chem Rev 113(8):6207–6233
Bahmyari S, Jamali Z, Khatami SH, Vakili O, Roozitalab M, Savardashtaki A et al (2021) microRNAs in female infertility: an overview. Cell Biochem Funct 39(8):955–969
Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15(6):321–333
Mafi A, Rahmati A, Babaei Aghdam Z, Salami R, Salami M, Vakili O et al (2022) Recent insights into the microRNA-dependent modulation of gliomas from pathogenesis to diagnosis and treatment. Cell Mol Biol Lett 27(1):1–32
Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003;17(24):3011-6
Vishnoi A, Rani S (2017) MiRNA Biogenesis and regulation of diseases: an overview. Methods Mol Biol 1509:1–10
Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D’Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY). 2016;8(4):603-619
Azimi Sanavi M, Mahdavian F, Dorosti N, Karami N, Karami S, Khatami SH et al A review of highly sensitive electrochemical genosensors for microRNA detection: a novel diagnostic platform for neurodegenerative diseases diagnostics. Biotechnol Appl Biochem
Lujambio A, Lowe SW (2012) The microcosmos of cancer. Nature 482(7385):347–355
Si W, Shen J, Zheng H, Fan W (2019) The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 11(1):25
Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM et al (2012) Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A 109(26):E1695–E1704
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E et al (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99(24):15524–15529
Ghasabi M, Mansoori B, Mohammadi A, Duijf PH, Shomali N, Shirafkan N et al (2019) MicroRNAs in cancer drug resistance: basic evidence and clinical applications. J Cell Physiol 234(3):2152–2168
Giovannetti E, Erozenci A, Smit J, Danesi R, Peters GJ (2012) Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit Rev Oncol Hematol 81(2):103–122
Liang YN, Tang YL, Ke ZY, Chen YQ, Luo XQ, Zhang H et al (2017) MiR-124 contributes to glucocorticoid resistance in acute lymphoblastic leukemia by promoting proliferation, inhibiting apoptosis and targeting the glucocorticoid receptor. J Steroid Biochem Mol Biol 172:62–68
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294(5543):858–862
Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ et al (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71(3):515–526
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477(7364):295–300
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22(9):1775–1789
Zhang X, Hong R, Chen W, Xu M, Wang L (2019) The role of long noncoding RNA in major human disease. Bioorg Chem 92:103214
Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22(2):96–118
Gourvest M, Brousset P, Bousquet M (2019) Long noncoding RNAs in acute myeloid leukemia: functional characterization and clinical relevance. Cancers (Basel) 11(11):1638
Novikova IV, Hennelly SP, Tung CS, Sanbonmatsu KY (2013) Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol 425(19):3731–3746
Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20(3):300–307
McCabe EM, Rasmussen TP (2021) lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol 75:38–48
Alessio E, Bonadio RS, Buson L, Chemello F, Cagnin S (2020) A single cell but many different transcripts: a journey into the world of long non-coding RNAs. Int J Mol Sci 21(1):302
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y et al (2019) HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell 36(6):645–59.e8
Li W, Zhai L, Wang H, Liu C, Zhang J, Chen W et al (2016) Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 7(19):27778–27786
Zhang XW, Bu P, Liu L, Zhang XZ, Li J (2015) Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun 462(3):227–232
Movahedpour A, Vakili O, Khalifeh M, Mousavi P, Mahmoodzadeh A, Taheri-Anganeh M et al (2022) Mammalian target of rapamycin (mTOR) signaling pathway and traumatic brain injury: a novel insight into targeted therapy. Cell Biochem Funct 40(3):232–247
Yu Y, Kou D, Liu B, Huang Y, Li S, Qi Y et al (2020) LncRNA MEG3 contributes to drug resistance in acute myeloid leukemia by positively regulating ALG9 through sponging miR-155. Int J Lab Hematol 42(4):464–472
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. Rna 20(12):1829–1842
Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T (2017) Recent progress in circular RNAs in human cancers. Cancer Lett 404:8–18
Salami R, Salami M, Mafi A, Vakili O, Asemi Z (2022) Circular RNAs and glioblastoma multiforme: focus on molecular mechanisms. Cell Commun Signal 20(1):13
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH et al (2013) Circular intronic long noncoding RNAs. Mol Cell 51(6):792–806
Mafi A, Yadegar N, Salami M, Salami R, Vakili O, Aghadavod E (2021) Circular RNAs; powerful microRNA sponges to overcome diabetic nephropathy. Pathol Res Pract 227:153618
Najafi S, Zarch SMA, Majidpoor J, Pordel S, Aghamiri S, Rasul MF et al (2022) Recent insights into the roles of circular RNAs in human brain development and neurologic diseases. Int J Biol Macromol 225:1038–1048
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X et al (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–264
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338
Vakili O, Asili P, Babaei Z, Mirahmad M, Keshavarzmotamed A, Asemi Z, et al (2022) Circular RNAs in Alzheimer’s disease: a new perspective of diagnostic and therapeutic targets. CNS & Neurological Disorders-Drug Targets (Formerly Current Drug Targets-CNS & Neurological Disorders). https://doi.org/10.2174/1871527321666220829164211
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M et al (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66
Dorostgou Z, Yadegar N, Dorostgou Z, Khorvash F, Vakili O (2022) Novel insights into the role of circular RNAs in Parkinson disease: an emerging renaissance in the management of neurodegenerative diseases. J Neurosci Res 100(9):1775–1790
Kristensen LS, Hansen TB, Venø MT, Kjems J (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37(5):555–565
Croce CM (2016) Retraction: are circRNAs involved in cancer pathogenesis? Nat Rev Clin Oncol 13(11):658
Liu J, Kong F, Lou S, Yang D, Gu L (2018) Global identification of circular RNAs in chronic myeloid leukemia reveals hsa_circ_0080145 regulates cell proliferation by sponging miR-29b. Biochem Biophys Res Commun 504(4):660–665
Xu T, Wang M, Jiang L, Ma L, Wan L, Chen Q et al (2020) CircRNAs in anticancer drug resistance: recent advances and future potential. Mol Cancer 19(1):127
Cao HX, Miao CF, Sang LN, Huang YM, Zhang R, Sun L et al (2020) Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci 243:117255
Liu Y, Dong Y, Zhao L, Su L, Luo J (2018) Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol 53(4):1752–1762
Vasan N, Baselga J, Hyman DM (2019) A view on drug resistance in cancer. Nature 575(7782):299–309
Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY et al (2009) Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells 27(9):2059–2068
Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M et al (2005) EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352(8):786–792
Ward RA, Fawell S, Floc'h N, Flemington V, McKerrecher D, Smith PD (2021) Challenges and opportunities in cancer drug resistance. Chem Rev 121(6):3297–3351
Moeinabadi-Bidgoli K, Rezaee M, Rismanchi H, Mohammadi MM, Babajani A (2022) Mesenchymal stem cell-derived antimicrobial peptides as potential anti-neoplastic agents: new insight into anticancer mechanisms of stem cells and exosomes. Front Cell Dev Biol 10:900418
Farawela HM, Khorshied MM, Kassem NM, Kassem HA, Zawam HM (2014) The clinical relevance and prognostic significance of adenosine triphosphate ATP-binding cassette (ABCB5) and multidrug resistance (MDR1) genes expression in acute leukemia: an Egyptian study. J Cancer Res Clin Oncol 140(8):1323–1330
Ji Q, Qiu L (2016) Mechanism study of PEGylated polyester and β-cyclodextrin integrated micelles on drug resistance reversal in MRP1-overexpressed HL60/ADR cells. Colloids Surf B Biointerfaces 144:203–213
Hart SM, Ganeshaguru K, Scheper RJ, Prentice HG, Hoffbrand AV, Mehta AB (1997) Expression of the human major vault protein LRP in acute myeloid leukemia. Exp Hematol 25(12):1227–1232
Shtil AA, Ktitorova OV, Kakpakova ES, Holian O (2000) Differential effects of the MDR1 (multidrug resistance) gene-activating agents on protein kinase C: evidence for redundancy of mechanisms of acquired MDR in leukemia cells. Leuk Lymphoma 40(1-2):191–195
Perl AE, Altman JK, Cortes J, Smith C, Litzow M, Baer MR et al (2017) Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: a multicentre, first-in-human, open-label, phase 1-2 study. Lancet Oncol 18(8):1061–1075
Stephen AG, Esposito D, Bagni RK, McCormick F (2014) Dragging ras back in the ring. Cancer Cell 25(3):272–281
Chen P, Huang H, Wu J, Lu R, Wu Y, Jiang X et al (2015) Bone marrow stromal cells protect acute myeloid leukemia cells from anti-CD44 therapy partly through regulating PI3K/Akt-p27(Kip1) axis. Mol Carcinog 54(12):1678–1685
Tazzari PL, Cappellini A, Ricci F, Evangelisti C, Papa V, Grafone T et al (2007) Multidrug resistance-associated protein 1 expression is under the control of the phosphoinositide 3 kinase/Akt signal transduction network in human acute myelogenous leukemia blasts. Leukemia 21(3):427–438
Liu X, Liao W, Peng H, Luo X, Luo Z, Jiang H et al (2016) miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J Cancer Res Clin Oncol 142(1):77–87
Tian P, Yan L (2016) Inhibition of microRNA-149-5p induces apoptosis of acute myeloid leukemia cell line THP-1 by targeting Fas ligand (FASLG). Med Sci Monit 22:5116–5123
Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S et al (1996) Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med 2(5):561–566
Milojkovic D, Apperley J (2009) Mechanisms of resistance to imatinib and second-generation tyrosine inhibitors in chronic myeloid leukemia. Clin Cancer Res 15(24):7519–7527
Eiring AM, Page BDG, Kraft IL, Mason CC, Vellore NA, Resetca D et al (2015) Combined STAT3 and BCR-ABL1 inhibition induces synthetic lethality in therapy-resistant chronic myeloid leukemia. Leukemia 29(3):586–597
Ma L, Shan Y, Bai R, Xue L, Eide CA, Ou J et al (2014) A therapeutically targetable mechanism of BCR-ABL-independent imatinib resistance in chronic myeloid leukemia. Sci Transl Med 6(252):252ra121
Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM et al (2004) Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med 351(6):533–542
Bhojwani D, Kang H, Moskowitz NP, Min DJ, Lee H, Potter JW et al (2006) Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a children’s oncology group study. Blood 108(2):711–717
Swerts K, De Moerloose B, Dhooge C, Laureys G, Benoit Y, Philippé J (2006) Prognostic significance of multidrug resistance-related proteins in childhood acute lymphoblastic leukaemia. Eur J Cancer 42(3):295–309
Pal Singh S, Dammeijer F, Hendriks RW (2018) Role of Bruton’s tyrosine kinase in B cells and malignancies. Mol Cancer 17(1):1–23
Liang C, Tian D, Ren X, Ding S, Jia M, Xin M et al (2018) The development of Bruton’s tyrosine kinase (BTK) inhibitors from 2012 to 2017: a mini-review. Eur J Med Chem 151:315–326
Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS et al (2014) Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med 370(24):2286–2294
Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA (2022) Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther 7(1):121
Wang WT, Han C, Sun YM, Chen TQ, Chen YQ (2019) Noncoding RNAs in cancer therapy resistance and targeted drug development. J Hematol Oncol. 12(1):55
Zebisch A, Hatzl S, Pichler M, Wölfler A, Sill H (2016) Therapeutic resistance in acute myeloid leukemia: the role of non-coding RNAs. Int J Mol Sci 17(12):2080
Taghvimi S, Vakili O, Soltani Fard E, Khatami SH, Karami N, Taheri-Anganeh M et al (2022) Exosomal microRNAs and long noncoding RNAs: novel mediators of drug resistance in lung cancer. J Cell Physiol 237(4):2095–2106
Pavlíková L, Šereš M, Breier A, Sulová Z (2022) The roles of microRNAs in cancer multidrug resistance. Cancers (Basel) 14(4):1090
Li H, Yang BB (2013) Friend or foe: the role of microRNA in chemotherapy resistance. Acta Pharmacol Sin 34(7):870–879
Sun YP, Lu F, Han XY, Ji M, Zhou Y, Zhang AM et al (2016) MiR-424 and miR-27a increase TRAIL sensitivity of acute myeloid leukemia by targeting PLAG1. Oncotarget 7(18):25276–25290
Liu Y, Lei P, Qiao H, Sun K, Lu X, Bao F et al (2020) MicroRNA-33b regulates sensitivity to daunorubicin in acute myelocytic leukemia by regulating eukaryotic translation initiation factor 5A-2. J Cell Biochem 121(1):385–393
Liu Y, Lei P, Qiao H, Sun K, Lu X, Bao F et al (2019) miR-9 Enhances the chemosensitivity of AML cells to daunorubicin by targeting the EIF5A2/MCL-1 axis. Int J Biol Sci 15(3):579–586
Vandewalle V, Essaghir A, Bollaert E, Lenglez S, Graux C, Schoemans H et al (2021) miR-15a-5p and miR-21-5p contribute to chemoresistance in cytogenetically normal acute myeloid leukaemia by targeting PDCD4, ARL2 and BTG2. J Cell Mol Med 25(1):575–585
Bai H, Xu R, Cao Z, Wei D, Wang C (2011) Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett 585(2):402–408
Li Y, Zhu X, Gu J, Dong D, Yao J, Lin C et al (2010) Anti-miR-21 oligonucleotide sensitizes leukemic K562 cells to arsenic trioxide by inducing apoptosis. Cancer Sci 101(4):948–954
Bollaert E, Claus M, Vandewalle V, Lenglez S, Essaghir A, Demoulin J-B et al (2021) MiR-15a-5p confers chemoresistance in acute myeloid leukemia by inhibiting autophagy induced by daunorubicin. Int J Mol Sci 22(10):5153
Wang Z, Fang Z, Lu R, Zhao H, Gong T, Liu D et al (2019) MicroRNA-204 potentiates the sensitivity of acute myeloid leukemia cells to arsenic trioxide. Oncol Res 27(9):1035
Ganesan S, Palani HK, Lakshmanan V, Balasundaram N, Alex AA, David S et al (2019) Stromal cells downregulate miR-23a-5p to activate protective autophagy in acute myeloid leukemia. Cell Death Dis 10(10):1–14
Zhang Y, Liu Y, Xu X (2017) Upregulation of miR-142-3p improves drug sensitivity of acute myelogenous leukemia through reducing P-glycoprotein and repressing autophagy by targeting HMGB1. Transl Oncol 10(3):410–418
Lu F, Zhang J, Ji M, Li P, Du Y, Wang H et al (2014) miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J Oncol 45(1):383–392
Zhang Y, Chu X, Wei Q (2021) MiR-451 promotes cell apoptosis and inhibits autophagy in pediatric acute myeloid leukemia by targeting HMGB1. J Environ Pathol Toxicol Oncol 40(2):45–53
Dai C-W, Bai Q-W, Zhang G-S, Cao Y-X, Shen J-K, Pei M-F et al (2014) MicroRNA let-7f is down-regulated in patients with refractory acute myeloid leukemia and is involved in chemotherapy resistance of adriamycin-resistant leukemic cells. Leuk Lymphoma 55(7):1645–1648
Cao YX, Wen F, Luo ZY, Long XX, Luo C, Liao P et al (2020) Downregulation of microRNA let-7f mediated the adriamycin resistance in leukemia cell line. J Cell Biochem 121(10):4022–4033
Krakowsky RH, Wurm AA, Gerloff D, Katzerke C, Bräuer-Hartmann D, Hartmann J-U et al (2018) miR-451a abrogates treatment resistance in FLT3-ITD-positive acute myeloid leukemia. Blood Cancer J 8(3):1–4
Jia Y, Liu W, Zhan H-E, Yi X-P, Liang H, Zheng Q-L et al (2020) Roles of hsa-miR-12462 and SLC9A1 in acute myeloid leukemia. J Hematol Oncol 13(1):101
Xiao Y, Deng T, Su C, Shang Z (2017) MicroRNA 217 inhibits cell proliferation and enhances chemosensitivity to doxorubicin in acute myeloid leukemia by targeting KRAS. Oncol Lett 13(6):4986–4994
Liu H, Liu M, Zhang J, Liang Y (2020) Downregulated miR-130a enhances the sensitivity of acute myeloid leukemia cells to adriamycin. Mol Med Rep 22(4):2810–2816
Li X, Xu L, Sheng X, Cai J, Liu J, Yin T et al (2018) Upregulated microRNA-146a expression induced by granulocyte colony-stimulating factor enhanced low-dosage chemotherapy response in aged acute myeloid leukemia patients. Exp Hematol 68:66–79.e3
L-j T, G-k S, T-j Z, Wu D-h, J-d Z, Ma B-b et al (2019) Down-regulation of miR-29c is a prognostic biomarker in acute myeloid leukemia and can reduce the sensitivity of leukemic cells to decitabine. Cancer Cell Int 19(1):177
Wu YY, Lai HF, Huang TC, Chen YG, Ye RH, Chang PY et al (2021) Aberrantly reduced expression of miR-342-5p contributes to CCND1-associated chronic myeloid leukemia progression and imatinib resistance. Cell Death Dis 12(10):908
Dong Y, Lin Y, Gao X, Zhao Y, Wan Z, Wang H et al (2019) Targeted blocking of miR328 lysosomal degradation with alkalized exosomes sensitizes the chronic leukemia cells to imatinib. Appl Microbiol Biotechnol 103(23-24):9569–9582
Jin J, Yao J, Yue F, Jin Z, Li D, Wang S (2018) Decreased expression of microRNA-214 contributes to imatinib mesylate resistance of chronic myeloid leukemia patients by upregulating ABCB1 gene expression. Exp Ther Med 16(3):1693–1700
Soltani I, Douzi K, Gharbi H, Benhassine I, Teber M, Amouri H et al (2017) Downregulation of miR-451 in Tunisian chronic myeloid leukemia patients: potential implication in imatinib resistance. Hematology 22(4):201–207
Li YL, Tang JM, Chen XY, Luo B, Liang GH, Qu Q et al (2020) MicroRNA-153-3p enhances the sensitivity of chronic myeloid leukemia cells to imatinib by inhibiting B-cell lymphoma-2-mediated autophagy. Hum Cell 33(3):610–618
Chen P-H, Liu A-J, Ho K-H, Chiu Y-T, Anne Lin Z-H, Lee Y-T et al (2018) microRNA-199a/b-5p enhance imatinib efficacy via repressing WNT2 signaling-mediated protective autophagy in imatinib-resistant chronic myeloid leukemia cells. Chem Biol Interact 291:144–151
Jiang X, Cheng Y, Hu C, Zhang A, Ren Y, Xu X (2019) MicroRNA-221 sensitizes chronic myeloid leukemia cells to imatinib by targeting STAT5. Leuk Lymphoma 60(7):1709–1720
Sun H, Li Y, Wang X, Zhou X, Rong S, Liang D et al (2022) TRIB2 regulates the expression of miR-33a-5p through the ERK/c-Fos pathway to affect the imatinib resistance of chronic myeloid leukemia cells. Int J Oncol 60(5):49
Deng Y, Li X, Feng J, Zhang X (2018) Overexpression of miR-202 resensitizes imatinib resistant chronic myeloid leukemia cells through targetting Hexokinase 2. Biosci Rep 38(3):BSR20171383
Farhadi E, Zaker F, Safa M, Rezvani MR (2016) miR-101 sensitizes K562 cell line to imatinib through Jak2 downregulation and inhibition of NF-κB target genes. Tumour Biol 37(10):14117–14128
Zhu X, Zhang J, Sun Y, Wang Y, Liu Q, Li P et al (2022) Restoration of miR-23a expression by chidamide sensitizes CML cells to imatinib treatment with concomitant downregulation of CRYAB. Bioengineered 13(4):8881–8892
Ramachandran SS, Muiwo P, Ahmad HM, Pandey RM, Singh S, Bakhshi S et al (2017) miR-505-5p and miR-193b-3p: potential biomarkers of imatinib response in patients with chronic myeloid leukemia. Leuk Lymphoma 58(8):1981–1984
Lin H, Rothe K, Chen M, Wu A, Babaian A, Yen R et al (2020) The miR-185/PAK6 axis predicts therapy response and regulates survival of drug-resistant leukemic stem cells in CML. Blood 136(5):596–609
Li Y, Luo S, Dong W, Song X, Zhou H, Zhao L et al (2016) Alpha-2, 3-sialyltransferases regulate the multidrug resistance of chronic myeloid leukemia through miR-4701-5p targeting ST3GAL1. Lab Invest 96(7):731–740
Zhou H, Li Y, Liu B, Shan Y, Li Y, Zhao L et al (2017) Downregulation of miR-224 and let-7i contribute to cell survival and chemoresistance in chronic myeloid leukemia cells by regulating ST3GAL IV expression. Gene 626:106–118
Ma J, Wu D, Yi J, Yi Y, Zhu X, Qiu H et al (2019) MiR-378 promoted cell proliferation and inhibited apoptosis by enhanced stem cell properties in chronic myeloid leukemia K562 cells. Biomed Pharmacother 112:108623
Nie ZY, Yao M, Yang Z, Yang L, Liu XJ, Yu J et al (2020) De-regulated STAT5A/miR-202-5p/USP15/Caspase-6 regulatory axis suppresses CML cell apoptosis and contributes to Imatinib resistance. J Exp Clin Cancer Res 39(1):17
Min QH, Wang XZ, Zhang J, Chen QG, Li SQ, Liu XQ et al (2018) Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Exp Cell Res 362(2):386–393
Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang H et al (2012) An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-α and cAMP/PKA pathways. Leukemia 26(4):769–777
Piatopoulou D, Avgeris M, Marmarinos A, Xagorari M, Baka M, Doganis D et al (2017) miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br J Cancer 117(6):801–812
Sheybani Z, Rahgozar S, Ghodousi ES (2019) The Hedgehog signal transducer smoothened and microRNA-326: pathogenesis and regulation of drug resistance in pediatric B-cell acute lymphoblastic leukemia. Cancer Manag Res 11:7621–7630
Zamani A, Fattahi Dolatabadi N, Houshmand M, Nabavizadeh N (2021) miR-324-3p and miR-508-5p expression levels could serve as potential diagnostic and multidrug-resistant biomarkers in childhood acute lymphoblastic leukemia. Leuk Res 109:106643
Qian L, Zhang W, Lei B, He A, Ye L, Li X et al (2016) MicroRNA-101 regulates T-cell acute lymphoblastic leukemia progression and chemotherapeutic sensitivity by targeting Notch1. Oncol Rep 36(5):2511–2516
Saleh LM, Wang W, Herman SE, Saba NS, Anastas V, Barber E et al (2017) Ibrutinib downregulates a subset of miRNA leading to upregulation of tumor suppressors and inhibition of cell proliferation in chronic lymphocytic leukemia. Leukemia 31(2):340–349
Zhu D-X, Zhu W, Fang C, Fan L, Zou Z-J, Wang Y-H et al (2012) miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 33(7):1294–1301
Pfeffer CM, Singh ATK (2018) Apoptosis: a target for anticancer therapy. Int J Mol Sci 19(2):448
Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B et al (2012) Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif 45(6):487–498
Ghavami S, Zamani M, Ahmadi M, Erfani M, Dastghaib S, Darbandi M et al (2022) Epigenetic regulation of autophagy in gastrointestinal cancers. Biochim Biophys Acta (BBA) - Mol Basis Dis 1868(11):166512
Khan KH, Blanco-Codesido M, Molife LR (2014) Cancer therapeutics: targeting the apoptotic pathway. Crit Rev Oncol Hematol 90(3):200–219
Singh R, Letai A, Sarosiek K (2019) Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol 20(3):175–193
Wei Y, Cao Y, Sun R, Cheng L, Xiong X, Jin X et al (2020) Targeting Bcl-2 proteins in acute myeloid leukemia. Front Oncol 10:584974
Trivedi R, Mishra DP (2015) Trailing TRAIL resistance: novel targets for TRAIL sensitization in cancer cells. Front Oncol 5:69
Dombret H, Gardin C (2016) An update of current treatments for adult acute myeloid leukemia. Blood 127(1):53–61
Daver N, Wei AH, Pollyea DA, Fathi AT, Vyas P, DiNardo CD (2020) New directions for emerging therapies in acute myeloid leukemia: the next chapter. Blood Cancer J 10(10):107
Murphy T, Yee KWL (2017) Cytarabine and daunorubicin for the treatment of acute myeloid leukemia. Expert Opin Pharmacother 18(16):1765–1780
Wang H, Guo M, Wei H, Chen Y (2021) Targeting MCL-1 in cancer: current status and perspectives. J Hematol Oncol 14(1):67
Low CG, Luk IS, Lin D, Fazli L, Yang K, Xu Y et al (2013) BIRC6 protein, an inhibitor of apoptosis: role in survival of human prostate cancer cells. PLoS One 8(2):e55837
Seo W, Silwal P, Song I-C, Jo E-K (2022) The dual role of autophagy in acute myeloid leukemia. J Hematol Oncol 15(1):51
Fukuda Y, Lian S, Schuetz JD (2015) Leukemia and ABC transporters. Adv Cancer Res 125:171–196
Zheng Y, Ma L, Sun Q (2021) Clinically-relevant ABC transporter for anti-cancer drug resistance. Front Pharmacol 12:648407
Wang F, Wang XS, Yang GH, Zhai PF, Xiao Z, Xia LY et al (2012) miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol Biol Rep 39(3):2713–2722
Chen R, Kang R, Tang D (2022) The mechanism of HMGB1 secretion and release. Exp Mol Med 54(2):91–102
Okabe S, Tauchi T, Katagiri S, Tanaka Y, Ohyashiki K (2014) Combination of the ABL kinase inhibitor imatinib with the Janus kinase 2 inhibitor TG101348 for targeting residual BCR-ABL-positive cells. J Hematol Oncol 7(1):37
Poudel G, Tolland MG, Hughes TP, Pagani IS (2022) Mechanisms of resistance and implications for treatment strategies in chronic myeloid leukaemia. Cancers 14(14):3300
Patel AB, O'Hare T, Deininger MW (2017) Mechanisms of resistance to ABL kinase inhibition in chronic myeloid leukemia and the development of next generation ABL kinase inhibitors. Hematol Oncol Clin North Am 31(4):589–612
Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M et al (2009) Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest 119(5):1109–1123
Alves R, Gonçalves AC, Rutella S, Almeida AM, De Las RJ, Trougakos IP et al (2021) Resistance to tyrosine kinase inhibitors in chronic myeloid leukemia-from molecular mechanisms to clinical relevance. Cancers (Basel) 13(19):4820
Ye D, Wolff N, Li L, Zhang S, Ilaria RL Jr (2006) STAT5 signaling is required for the efficient induction and maintenance of CML in mice. Blood 107(12):4917–4925
Li J, Gao N, Gao Z, Liu W, Pang B, Dong X et al (2021) The emerging role of exosomes in cancer chemoresistance. Front Cell Dev Biol 9:737962
Guo QR, Wang H, Yan YD, Liu Y, Su CY, Chen HB et al (2020) The role of exosomal microRNA in cancer drug resistance. Front Oncol 10:472
Ghodousi ES, Rahgozar S (2018) MicroRNA-326 and microRNA-200c: Two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem 119(7):6024–6032
Zhang H, Sun Z, Liu Z, Song C (2018) Overcoming the emerging drug resistance of smoothened: an overview of small-molecule SMO antagonists with antiresistance activity. Future Med Chem 10(24):2855–2875
Wang A, Chen Y, Shi L, Li M, Li L, Wang S et al (2022) Tumor-suppressive MEG3 induces microRNA-493-5p expression to reduce arabinocytosine chemoresistance of acute myeloid leukemia cells by downregulating the METTL3/MYC axis. J Transl Med 20(1):288
Yan J, Yao L, Li P, Wu G, Lv X (2022) Long non-coding RNA MIR17HG sponges microRNA-21 to upregulate PTEN and regulate homoharringtonine-based chemoresistance of acute myeloid leukemia cells. Oncol Lett 23(1):24
Dong X, Fang Z, Yu M, Zhang L, Xiao R, Li X et al (2018) Knockdown of long noncoding RNA HOXA-AS2 suppresses chemoresistance of acute myeloid leukemia via the miR-520c-3p/S100A4 Axis. Cell Physiol Biochem 51(2):886–896
Sun H, Sun Y, Chen Q, Xu Z (2020) LncRNA KCNQ1OT1 contributes to the progression and chemoresistance in acute myeloid leukemia by modulating Tspan3 through suppressing miR-193a-3p. Life Sci 241:117161
Kang Y, Zhang S, Cao W, Wan D, Sun L (2020) Knockdown of LncRNA CRNDE suppresses proliferation and P-glycoprotein-mediated multidrug resistance in acute myelocytic leukemia through the Wnt/β-catenin pathway. Biosci Rep 40:6
Chen L, Zhao H, Wang C, Hu N (2019) TUG1 knockdown enhances adriamycin cytotoxicity by inhibiting glycolysis in adriamycin-resistant acute myeloid leukemia HL60/ADR cells. RSC Adv. 9(19):10897–10904
Zhang B, Sun YF, Zhang XM, Jiang N, Chen Q (2020) TUG1 weakens the sensitivity of acute myeloid leukemia cells to cytarabine by regulating miR-655-3p/CCND1 axis. Eur Rev Med Pharmacol Sci 24(9):4940–4953
Li J, Wang M, Chen X (2020) Long non-coding RNA UCA1 modulates cell proliferation and apoptosis by regulating miR-296-3p/Myc axis in acute myeloid leukemia. Cell Cycle 19(12):1454–1465
Chen L, Hu N, Wang C, Zhao H (2020) HOTAIRM1 knockdown enhances cytarabine-induced cytotoxicity by suppression of glycolysis through the Wnt/β-catenin/PFKP pathway in acute myeloid leukemia cells. Arch Biochem Biophys 680:108244
Liang L, Gu W, Li M, Gao R, Zhang X, Guo C et al (2021) The long noncoding RNA HOTAIRM1 controlled by AML1 enhances glucocorticoid resistance by activating RHOA/ROCK1 pathway through suppressing ARHGAP18. Cell Death Dis 12(7):702
Liu JM, Li M, Luo W, Sun HB (2021) Curcumin attenuates adriamycin-resistance of acute myeloid leukemia by inhibiting the lncRNA HOTAIR/miR-20a-5p/WT1 axis. Lab Invest 101(10):1308–1317
Zhang H, Zhao Y, Liu X, Liu Y, Wang X, Fu Y et al (2021) A novel upregulated LncRNA-AC026150.8 promotes chemo-resistance and predicts poor prognosis in acute myeloid leukemia. Cancer Med 10(23):8614–8629
Cui C, Wang Y, Gong W, He H, Zhang H, Shi W et al (2021) Long non-coding RNA LINC00152 regulates self-renewal of leukemia stem cells and induces chemo-resistance in acute myeloid leukemia. Front Oncol 11:694021
Zhou H, Jia X, Yang F, Shi P (2021) Long noncoding RNA SATB1-AS1 contributes to the chemotherapy resistance through the microRNA-580/ 2′-5′-oligoadenylate synthetase 2 axis in acute myeloid leukemia. Bioengineered 12(1):6403–6417
Zhang H, Liu L, Chen L, Liu H, Ren S, Tao Y (2021) Long noncoding RNA DANCR confers cytarabine resistance in acute myeloid leukemia by activating autophagy via the miR-874-3P/ATG16L1 axis. Mol Oncol 15(4):1203–1216
Yang Y, Dai W, Sun Y, Zhao Z (2019) Long non-coding RNA linc00239 promotes malignant behaviors and chemoresistance against doxorubicin partially via activation of the PI3K/Akt/mTOR pathway in acute myeloid leukaemia cells. Oncol Rep 41(4):2311–2320
Wang C, Li L, Li M, Wang W, Liu Y, Wang S (2020) Silencing long non-coding RNA XIST suppresses drug resistance in acute myeloid leukemia through down-regulation of MYC by elevating microRNA-29a expression. Mol Med 26(1):114
Hu N, Chen L, Wang C, Zhao H (2019) MALAT1 knockdown inhibits proliferation and enhances cytarabine chemosensitivity by upregulating miR-96 in acute myeloid leukemia cells. Biomed Pharmacother 112:108720
Xue L, Li C, Ren J, Wang Y (2021) KDM4C contributes to cytarabine resistance in acute myeloid leukemia via regulating the miR-328-3p/CCND2 axis through MALAT1. Ther Adv Chronic Dis 12:2040622321997259
Li Q, Wang J (2019) Long noncoding RNA ZFAS1 enhances adriamycin resistance in pediatric acute myeloid leukemia through the miR-195/Myb axis. RSC Adv 9(48):28126–28134
Wang D, Zeng T, Lin Z, Yan L, Wang F, Tang L et al (2020) Long non-coding RNA SNHG5 regulates chemotherapy resistance through the miR-32/DNAJB9 axis in acute myeloid leukemia. Biomed Pharmacother 123:109802
Luo J, Gao Y, Lin X, Guan X (2021) Systematic analysis reveals a lncRNA-miRNA-mRNA network associated with dasatinib resistance in chronic myeloid leukemia. Ann Palliat Med 10(2):1727–1738
Wen F, Cao YX, Luo ZY, Liao P, Lu ZW (2018) LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem Biophys Res Commun 507(1-4):1–8
Han Y, Ma Z (2021) LncRNA highly upregulated in liver cancer regulates imatinib resistance in chronic myeloid leukemia via the miR-150-5p/MCL1 axis. Anticancer Drugs 32(4):427–436
Shehata AMF, Gohar SF, Muharram NM, Eldin SMK (2022) LncRNA CCAT2 expression at diagnosis predicts imatinib response in chronic phase chronic myeloid leukemia patients. Leuk Res 116:106838
Liu J, Yang L, Liu X, Liu L, Liu M, Feng X et al (2022) lncRNA HOTTIP Recruits EZH2 to inhibit PTEN expression and participates in IM resistance in chronic myeloid leukemia. Stem Cells Int 2022:9993393
Dai H, Wang J, Huang Z, Zhang H, Wang X, Li Q et al (2021) LncRNA OIP5-AS1 promotes the autophagy-related imatinib resistance in chronic myeloid leukemia cells by regulating miR-30e-5p/ATG12 Axis. Technol Cancer Res Treat 20:15330338211052150
Liu Y, Li H, Zhao Y, Li D, Zhang Q, Fu J et al (2022) Knockdown of ADORA2A antisense RNA 1 inhibits cell proliferation and enhances imatinib sensitivity in chronic myeloid leukemia. Bioengineered 13(2):2296–2307
Xiao Y, Jiao C, Lin Y, Chen M, Zhang J, Wang J et al (2017) lncRNA UCA1 contributes to imatinib resistance by acting as a ceRNA against miR-16 in chronic myeloid leukemia cells. DNA Cell Biol 36(1):18–25
He B, Bai Y, Kang W, Zhang X, Jiang X (2017) LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res 7(8):1704–1713
Zhou X, Yuan P, Liu Q, Liu Z (2017) LncRNA MEG3 regulates imatinib resistance in chronic myeloid leukemia via suppressing microRNA-21. Biomol Ther (Seoul) 25(5):490–496
Zhang F, Ni H, Li X, Liu H, Xi T, Zheng L (2019) LncRNA FENDRR attenuates adriamycin resistance via suppressing MDR1 expression through sponging HuR and miR-184 in chronic myelogenous leukaemia cells. FEBS Lett 593(15):1993–2007
Ouimet M, Drouin S, Lajoie M, Caron M, St-Onge P, Gioia R et al (2016) A childhood acute lymphoblastic leukemia-specific lncRNA implicated in prednisolone resistance, cell proliferation, and migration. Oncotarget 8(5):7477–7488
Zhao Q, Zhao S, Li J, Zhang H, Qian C, Wang H et al (2019) TCF7L2 activated HOXA-AS2 decreased the glucocorticoid sensitivity in acute lymphoblastic leukemia through regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway. Biomed Pharmacother 109:1640–1649
Miller CR, Ruppert AS, Fobare S, Chen TL, Liu C, Lehman A et al (2017) The long noncoding RNA, treRNA, decreases DNA damage and is associated with poor response to chemotherapy in chronic lymphocytic leukemia. Oncotarget 8(16):25942–25954
Martelli AM, Nyåkern M, Tabellini G, Bortul R, Tazzari PL, Evangelisti C et al (2006) Phosphoinositide 3-kinase/Akt signaling pathway and its therapeutical implications for human acute myeloid leukemia. Leukemia 20(6):911–928
Li S, Yao W, Liu R, Gao L, Lu Y, Zhang H et al (2022) Long non-coding RNA LINC00152 in cancer: roles, mechanisms, and chemotherapy and radiotherapy resistance. Front Oncol 12:960193
Ding J, Zhang X, Xue J, Fang L, Ban C, Song B et al (2021) CircNPM1 strengthens adriamycin resistance in acute myeloid leukemia by mediating the miR-345-5p/FZD5 pathway. Cent Eur J Immunol 46(2):162–182
Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ, Wu WB (2019) CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol 70:42–54.e3
Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen ZZ et al (2019) CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res 85:106198
Wang J, Liang Y, Qin Y, Jiang G, Peng Y, Feng W (2022) circCRKL, a circRNA derived from CRKL, regulates BCR-ABL via sponging miR-877-5p to promote chronic myeloid leukemia cell proliferation. J Transl Med 20(1):395
Lu YH, Huang ZY (2021) Global identification of circular RNAs in imatinib (IM) resistance of chronic myeloid leukemia (CML) by modulating signaling pathways of circ_0080145/miR-203/ABL1 and circ 0051886/miR-637/ABL1. Mol Med 27(1):148
Zhong AN, Yin Y, Tang BJ, Chen L, Shen HW, Tan ZP et al (2021) CircRNA microarray profiling reveals hsa_circ_0058493 as a novel biomarker for imatinib-resistant CML. Front Pharmacol 12:728916
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z (2019) High circ_100053 predicts a poor outcome for chronic myeloid leukemia and is involved in imatinib resistance. Oncol Res. https://doi.org/10.3727/096504018X15412701483326
Ji W, Sun B, Su C (2017) Targeting microRNAs in cancer gene therapy. Genes 8(1):21
Ors-Kumoglu G, Gulce-Iz S, Biray-Avci C (2019) Therapeutic microRNAs in human cancer. Cytotechnology 71:411–425
Naidu S, Magee P, Garofalo M (2015) MiRNA-based therapeutic intervention of cancer. J Hematol Oncol 8:1–8
Rabaan AA, AlSaihati H, Bukhamsin R, Bakhrebah MA, Nassar MS, Alsaleh AA et al (2023) Application of CRISPR/Cas9 technology in cancer treatment: a future direction. Curr Oncol 30(2):1954–1976
Zhang H, Qin C, An C, Zheng X, Wen S, Chen W et al (2021) Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer 20:1–22
Chen M, Mao A, Xu M, Weng Q, Mao J, Ji J (2019) CRISPR-Cas9 for cancer therapy: Opportunities and challenges. Cancer Lett 447:48–55
Khan MI, Hossain MI, Hossain MK, Rubel M, Hossain K, Mahfuz A et al (2022) Recent progress in nanostructured smart drug delivery systems for cancer therapy: a review. ACS Appl Bio Mater 5(3):971–1012
Author information
Authors and Affiliations
Contributions
The authors confirm contributions to this paper as follows: AR: wrote the manuscript and prepared the figures and tables. AM: conceptualized the review and modified the manuscript. OV, FS, ZA, SY, YG, and MR: collected the relevant literature and proposed amendments. AS: critically revised and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rahmati, A., Mafi, A., Vakili, O. et al. Non-coding RNAs in leukemia drug resistance: new perspectives on molecular mechanisms and signaling pathways. Ann Hematol 103, 1455–1482 (2024). https://doi.org/10.1007/s00277-023-05383-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-023-05383-3