Abstract
In biodesulfurization (BD) at haloalkaline and dO2-limited conditions, sulfide-oxidizing bacteria (SOB) effectively convert sulfide into elemental sulfur that can be used in agriculture as a fertilizer and fungicide. Here we show which bacteria are present in this biotechnological process. 16S rRNA gene amplicon sequencing of biomass from ten reactors sampled in 2018 indicated the presence of 444 bacterial Amplicon Sequence Variants (ASVs). A core microbiome represented by 30 ASVs was found in all ten reactors, with Thioalkalivibrio sulfidiphilus as the most dominant species. The majority of these ASVs are phylogenetically related to bacteria previously identified in haloalkaline BD processes and in natural haloalkaline ecosystems. The source and composition of the feed gas had a great impact on the microbial community composition followed by alkalinity, sulfate, and thiosulfate concentrations. The halophilic SOB of the genus Guyparkeria (formerly known as Halothiobacillus) and heterotrophic SOB of the genus Halomonas were identified as potential indicator organisms of sulfate and thiosulfate accumulation in the BD process.
Key points
• Biodesulfurization (BD) reactors share a core microbiome
• The source and composition of the feed gas affects the microbial composition in the BD reactors
• Guyparkeria and Halomonas indicate high concentrations of sulfate and thiosulfate in the BD process
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hydrogen sulfide (H2S) is a malodorous, toxic, and corrosive gas, which can be harmful to all forms of life. This gas is produced in nature by sulfate-reducing bacteria found in anaerobic habitats, such as marine and freshwater sediments, as well as man-made processes (Muyzer and Stams 2008). Naturally, geothermic activity also releases H2S in the atmosphere. Nowadays, the petrochemical industry is the largest anthropogenic producer of SO2, produced upon combustion of H2S. The other sources include paper mills, the textile industry, biogas plants, landfills, and the rendering industry (Mahmood et al. 2007; Janssen et al. 2009). Conventional methods to remove H2S on-site include physical and chemical processes. Alternatively, more environmentally friendly and cost-effective haloalkaline BD processes employ haloalkaliphilic SOB to convert sulfide into elemental sulfur at haloalkaline conditions and low redox potential (Janssen et al. 2001). In these processes, H2S is first absorbed into an alkaline solution as a predominantly soluble bisulfide (HS−). The HS−-loaded solution is subsequently directed to a bioreactor whereby SOB oxidizes it to elemental sulfur at a low redox potential of around −300 mV (Sorokin et al. 2008; Janssen et al. 2009). The produced sulfur is harvested from a settler and can be further reused as fertilizer or fungicide. With this BD process, 99.84% of sulfide can be removed (de Rink et al. 2020). Besides sulfur, thiosulfate and sulfate are formed as side products through chemical oxidation of polysulfide and complete oxidation of reduced sulfur compounds mediated by SOB, respectively. Both compounds are undesirable, as their formation (i) increases acidification and consequently, the need for extra caustic supply, (ii) increases the demand of oxygen supply (= energy consumption), and (iii) decreases the sulfur selectivity of the process (van den Bosch et al. 2007). The rate of formation and removal of sulfate and thiosulfate from the process determines the concentration of these compounds in the reactor system and can be regarded as indicators of a less efficient process. We hypothesize that these indicators are directly linked to the activity of particular bacteria in the reactor.
Aerobic sulfur oxidation is largely performed by chemolithoautotrophic SOB along with facultative autotrophs, lithoheterotrophs, and obligate heterotrophs. The chemolithoautotrophic SOB possesses the ability to conserve energy from the oxidation of reduced sulfur compounds, such as sulfide, polysulfides, sulfur, thiosulfate, and polythionates with oxygen or nitrate as electron acceptor (Muyzer et al. 2013). They are members of four genera of haloalkaliphilic Gammaproteobacteria: Thioalkalimicrobium reclassified as Thiomicrospira (Boden et al. 2017), Thioalkalispira, Thioalkalivibrio, and Thioalkalibacter (Sorokin et al. 2013, 2015). The haloalkaliphilic Thiomicrospira and Thioalkalispira species can grow with thiosulfate or sulfide forming mostly sulfate as a product but can produce elemental sulfur under oxygen-limiting conditions (Sorokin et al. 2001; 2002). The genus Thioalkalivibrio includes multiple isolates (more than a hundred) from natural soda lakes and haloalkaline bioreactors that can oxidize various reduced sulfur compounds to sulfur or sulfate using oxygen or nitrate as an electron acceptor (Berben et al. 2019). Until today, ten species of Thioalkalivibrio have been well-characterized and a variety of other species have been identified based on their genomes (Ahn et al. 2017; Berben et al. 2019). Other SOBs from haloalkaline habitats are members belonging to the Alkalispirillum–Alkalilimnicola group, i.e., Alkalilimnicola halodurans, Alkalilimnicola ehrlichii MLHE-1 (Hoeft et al. 2007), Alkalispirillum mobile (Rijkenberg et al. 2001), and several unclassified members. Members of the Alkalispirillum–Alkalilimnicola group are facultatively autotrophic genera that can utilize CO as an additional e-donor (Sorokin et al. 2006). Other bacteria that might be present in the haloalkaline bioreactors are members of the genus Halomonas, which are heterotrophic bacteria that can oxidize thiosulfate to tetrathionate, which, in turn, can chemically oxidize sulfide to sulfur with the regeneration of thiosulfate, thus, potentially contributing to the overall sulfide oxidation to sulfur (Sorokin, 2003; Sorokin et al. 2008).
Multiple 16S rRNA sequence analyses of bacterial communities from some full- and pilot-scale BD processes has revealed the presence of both chemolithoautotrophic and heterotrophic SOB belonging to the Gammaproteobacteria, including Thioalkalivibrio, Thiomicrospira, and Halomonas species (de Graaff et al. 2011; Sorokin et al. 2012; Roman et al., 2016; Kiragosyan et al. 2019). Recently, the BD process has been improved by the addition of an anoxic reactor in series (Klok et al. 2017, 2018). In this BD concept, SOB was exposed to alternating conditions of subsequent anaerobic-sulfidic and microaerobic conditions. In the new process, line-up selectivity for sulfur formation of 96.5% was achieved, which considerably decreases NaOH consumption and bleed stream formation compared to the line-up without the anoxic reactor. Correspondingly the dominance of the facultative chemolithoautotrophic SOB Alkalilimnicola also increased in the novel BD process (de Rink et al. 2019). Although the aforementioned studies provide insight into the microbial community composition of some BD processes, detailed comparative information on the microbial community composition in different full-scale plants is still lacking. Besides, the presence of a possible core community in the BD process has not been studied before.
In this paper, we analyzed the microbial communities from eight full- and two pilot-scale biodesulfurization reactors along with a variety of physicochemical parameters. We determined the microbial diversity in these bioreactors and defined the presence of a core community. Moreover, we revealed a relation between the presence of specific community members at particular environmental conditions, such as alkalinity. We also showed that there are certain bacteria that can be linked to an increase in the undesired production of thiosulfate and sulfate.
Materials and methods
Description and sampling of full-and pilot-scale BD reactors
To determine the microbial community composition of BD reactors comprehensively, process solutions of eight full-scale BD reactors and two runs of a pilot-scale BD reactor were analyzed in 2018. Full-scale BD reactors studied were from industries that treated feed gas from different origins (Table 1). The pilot-scale BD reactors consisted of an extra reactor, namely an anoxic reactor in front of the oxic reactor. The difference between the pilot-scale and full-scale was the composition of the feed gas; for the pilot scale BD reactors, a synthetic gas with no contaminants was used, while the full-scale plants have organic sulfur compounds and hydrocarbons in the feed (Table 1). The process solution was collected from full-scale bioreactors sampling points. The solution was used to analyze the physical-chemical parameters and to extract genomic DNA for amplicon sequencing. For the pilot-scale BD reactors, the samples were collected from the sludge preserved at 4 °C after the process operation.
Physicochemical analyses of the bioreactor samples
To investigate the physical-chemical parameters of the collected samples, the samples were processed in the laboratory as early as possible. Prior to the analysis, the samples were kept at 4 °C and were filtered over a 0.45 µm membrane filter (HPF Millex, Merck, the Netherlands). Thiosulfate, sulfate, and sodium were measured by ion chromatography as described by Roman et al. (2015). Total inorganic carbon was quantified using a TOC-VCPH/CPN analyzer (Shimadzu, The Netherlands). Carbonate and bicarbonate ions concentration were estimated from the total inorganic carbon using the Henderson-Hasselbalch equation (Po and Senozan 2001) and further used to express the alkalinity of the system as concentration NaHCO3 (de Rink et al. 2019). The pH and conductivity of the well-mixed unfiltered samples were measured using the kit from Mettler Toledo, USA.
DNA extraction and 16S rRNA gene amplicon sequencing
For the microbial community composition analysis, genomic DNA was extracted from samples collected from the BD reactors in triplicate from each bioreactor sample. Twenty milliliters of sludge was centrifuged at 10,000 × g for 10 min at room temperature. The supernatant was discarded, and the pellet was suspended in a buffer containing 1M NaCl and phosphate-buffered-saline (PBS) to maintain the osmotic balance of the cells. The biomass was centrifuged again, and the obtained mixture of biomass and sulfur was used for the DNA extraction. The DNA was extracted using DNeasy PowerLyzer PowerSoil Kit (Qiagen, Germany) following the manufacturer’s instructions. The DNA was purified with DNA Clean & Concentrator-5 kit (Zymogen, USA). The integrity of the extracted genomic DNA was visualized by gel electrophoresis. PCR with 16S rRNA gene primers 27F and 1492R was performed, and amplicons were visualized by gel electrophoresis to verify the relative intensity of the amplicons. The extracted DNA was quantified using the QuantiFluor dsDNA system on a Quantus fluorometer (Promega, USA). Samples were diluted to a final concentration of 30 ng/µL of DNA and sent for Illumina MiSeq sequencing performed at commercial sequencing company MR DNA (www.mrdnalab.com, Shallowater, TX, USA). At the facility, samples were first amplified in triplicate using primers 515f (5ʹ-GTGYCAGCMGCCGCGGTAA-3ʹ) and 926r (5ʹ-CCGYCAATTYMTTTRAGTTT-3ʹ) with a bar-coded forward primer. This primer set covers the 16S rRNA gene V4–V5 variable region (Parada et al. 2016). Amplification was done three times using the HotStarTaq Plus Master Mix Kit (Qiagen, USA) under the following conditions: 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 53 °C for 40 s, and 72 °C for 1 min, with a final elongation step at 72 °C for 5 min. After amplification, the PCR products of three separate reactions of each sample were pooled, and PCR products were checked on a 2% (w/v) agarose gel to determine the success of amplification and the relative intensity of the bands. Then, multiple samples were pooled together in equimolar amounts based on their molecular weight and DNA concentrations. Pooled samples were purified using calibrated Ampure XP beads (Beckman Coulter, Indianapolis, IN, USA) and used for sequencing on a MiSeq system following the manufacturer’s guidelines. The sequence data has been submitted to the ENA database under study number PRJEB43104.
Amplicon sequence analysis
The 2 × 300 bp paired-end reads were generated by Illumina MiSeq sequencing. The reads generated were converted to paired-end format from a mixed paired format using the fastq processor software provided by MR DNA which removes the barcodes, primers and corrects the orientation of the reads. Microbiome analysis was performed with QIIME-2 version 2020.2 (Bolyen et al. 2019). Raw sequence data were demultiplexed and quality filtered using the q2‐demux plugin followed by denoising with DADA2 (Callahan et al. 2016) (via q2‐dada2) to remove the errors and chimeric sequences and identify all observed amplicon sequence variants (ASVs) (100% operational taxonomic units). All amplicon sequence variants (ASVs) were aligned with MAFFT (Katoh and Standley 2013) (via q2‐alignment) and used to construct a phylogeny tree with fasttree2 (Price et al. 2010) (via q2‐phylogeny). The taxonomy was assigned to the ASVs using the q2‐feature‐classifier (Bokulich et al. 2018), the classify-sklearn naive Bayes taxonomy classifier (Pedregosa et al. 2011) trained on nearly complete 16S rRNA sequences from the SILVA database version 138 (Quast et al. 2013; Yilmaz et al. 2014).
Alpha‐diversity metrics (observed OTUs and Shannon), beta diversity metrics (Bray‐Curtis dissimilarity), and Principal Coordinate Analysis (PCoA) were estimated using q2‐diversity after samples were rarefied (subsampled without replacement) to 33,340 sequences per sample.
Identification and phylogenetic analysis of the core community members
The core ASVs were identified using the QIIME 2 feature table core features plugin. To create a visual diagram depicting the core community, the ASV table was filtered based on industry type to generate five ASV tables specific to each industry type using QIIME 2 feature table filter samples plugin. The core ASVs for each industry type were identified using the QIIME 2 feature table core features plugin. The counts of core ASVs were used to create a Venn diagram to show the shared core community of all the BD reactors. The Venn diagram was created using the “InteractiVenn” tool (Heberle et al. 2015) whereby, each industry type is represented as one set. Next, the core ASV table for the whole data set was created using QIIME 2 feature table core features plugin. The representative sequences were filtered based on the core ASV tables using QIIME 2 filter sequences tool. The identified core sequences were aligned in SINA alignment (Pruesse et al. 2012) imported into ARB (Quast et al. 2013; Yilmaz et al. 2014) to create a phylogenetic tree showing the core ASVs and their close relatives.
Prediction of the functional potential of the core microbiome
The functional potential of the core community was predicted using the software program Tax4Fun2 that can predict the functional capabilities of the microbial communities based on 16S rRNA datasets (Wemheuer et al. 2018). The genomes of bacteria that were closely affiliated to the core community members were also included in the analyses as described in the pipeline (https://github.com/bwemheu/Tax4Fun2).
Co-occurrence network analysis
Co-occurrence networks were calculated with the software program GenePiper (Tong and Chan 2020) using the raw read counts as abundance type. In addition, we used the person correlation method; the correlation filter was set to 0.8 and the P-value to 0.05, and agglomeration was performed at the genus level.
Statistical analysis of the data
To identify the difference in microbial community composition between samples and different industry types, PCoA analysis was performed using the Bray Curtis similarity distances. Redundancy Analysis (RDA) was performed to determine which parameters among pH, salinity, alkalinity, conductivity, H2S/CO2 ratio, and concentrations of thiosulfate and sulfate significantly explain variation in microbial community composition. The analysis was performed in an R software environment (version 3.6.3) using the vegan package. The ASV table, tree, and taxonomy files created in QIIME 2 were imported in R for this analysis. As a prerequisite to RDA, the species frequency is converted to abundance data. Variance inflation factors were analyzed for all parameters to confirm the non-collinearity using package car (version 3.0-8). The significance of explanatory variables was analyzed using ANOVA-like analysis with 999 permutations, and a tri-plot was generated using the ggplot2 package in R.
In addition to the relationship, we were also interested in the differential abundance of ASVs/taxa with changing concentrations of sulfate, thiosulfate, and alkalinity. The differentials were generated using the q2-songbird plugin in QIIME 2 (Morton et al. 2019) by fitting environmental parameters to the multinomial regression model. The good fit was determined using the factor Q2. A value closer to 1 indicates a good fit and therefore represents the significant differential abundance of species due to change in parameter values. A value closer to 0 indicates a poor fit and no conclusion can be inferred from the model.
Results
Physicochemical analyses
Several physicochemical parameters of the full-scale BD-processes, such as pH, salinity, conductivity, alkalinity, and concentrations of sulfate and thiosulfate, were determined and are presented in Table 1. The BD-reactor treating biogas from the rendering industry had the highest measured sulfate and thiosulfate concentrations and the lowest alkalinity. In contrast, the BD-reactor that was treating gas from a landfill had low sulfate and thiosulfate concentrations but high alkalinity. In the BD processes that were treating gas from the petrochemical industry, only sulfate was detected. For all samples, the salinity varied between 0.8 and 1 M Na+, and the conductivity was between 40 and 78 mS/cm. The alkalinity ranged from 0.18 to 0.71 M and the pH from 8.1 to 9, indicating haloalkaline conditions in all reactors. The oxygen supply in all plants was controlled using ORP (van den Bosch et al. 2007). The measured range of the ORP at the installation ranged from −389 to −283 mV.
16S rRNA gene-based amplicon sequencing and microbial diversity analyses
Demultiplexing and denoising of the raw Illumina MiSeq sequences from the 30 biomass samples of the BD-reactors resulted in 33,342 to 73,228 reads and a total of 444 ASVs were assigned (Supplementary Table S1). Alpha diversity, which is indicative of the mean species diversity, based on both observed ASVs and Shannon indices varied significantly among the different BD reactors (Supplementary Fig. S1, Kruskal–Wallis, H = 17.81, p = 0.03 for observed ASVs and H = 27.64, p = 0.001 for Shannon).
The beta diversity, indicative of the difference between microbial communities from different samples, was significantly different among the samples (Bray Curtis distance; PERMANOVA: F = 13.9778, p = 0.001, permutations = 999). The Bray Curtis dissimilarity distances based on PCoA analysis (Fig. 1) showed great similarity in the microbial community composition of reactor samples from the paper mill industry and landfill gas treating plants, while the community composition of plants from the rendering industry and the petrochemical industry were more distinct. The BD reactors with no contaminants such as hydrocarbons and organic sulfur compounds in the feed gas had a less diverse community as compared to the BD reactors with a larger organic matter input. This was also observed for the two pilot plants that had an additional anoxic reactor. Two BD reactors employed in Oil and Gas industries had different contaminants in their feed gas, such as BTEX and thiols (Table 1), and had a different microbial community composition. The BD reactor of the rendering industry had high ammonia content and a very different microbial community composition than the other BD-reactors. The H2S/CO2 ratio in the feed gas and sulfur separation method did not have much effect on the microbial diversity among different BD reactors (Supplementary Fig. S2).
Principal coordinate analysis (PCoA) of Bray Curtis dissimilarity distances of the microbial communities of biodesulfurization reactors from different industries (see Table 1)
Identification of a core microbial community in the BD reactors
The core community of the bioreactor samples was defined based on the ASVs that were present in all BD-reactors. A total of 30 ASVs were present in all samples indicative of the “core microbial community.” The Venn diagram shown in Figure 2A indicates the distribution of all 444 ASVs among the samples grouped based on the industrial waste they treat and the 30 ASVs that are common to all BD-reactors studied. Interestingly these 30 ASVs together account for 72 to 90% of total microbial diversity in BD reactors (Fig. 2B).
Core community members found in the biodesulfurization reactors. A Venn diagram showing the number of unique and common ASVs. Sets represent the industry type the plants treat. The total count of each set is the number of ASVs commonly occurring in the reactors treating that industrial effluent. B Bar graph showing the average relative abundance of the 30 core ASVs in three replicates of each sample
Taxonomy-based analyses
Taxa assignment to identify ASVs revealed that microbial community members from the class Gammaproteobacteria had the highest relative abundance. These included the genera Thioalkalivibrio, Alkalilimnicola, Guyparkeria, Halomonas, Alkalispirillum, Vibrio, and Thiomicrospira.
The core microbial community with 30 ASVs consisted of 16 members from the class Gammaproteobacteria with known SOBs Thioalkalivibrio sulfidiphilus, Thioalkalivibrio halophilus, Thioalkalivibrio sp. K90mix, Alkalilimnicola ehrlichii, Halomonas, Thiomicrospira, and Guyparkeria along with Nitrincola and Wenzhouxiangella. Four members belonged to the class Alphaproteobacteria that includes members of genera Roseinatronobacter (a known haloalkaliphilic lithoheterotrophic SOB), Stappia, and one unclassified member of the family Rhodobacteraceae. The other four belonged to class Bacteroidia that includes members of genera Planktosalinus, Psychroflexus, Lentimicrobium, and Blvii28_wastewater-sludge_group. The next three members belonged to class Bacillia which includes members of genera Acholeplasma and Izimaplasma. The latter was also present among the abundant metagenomes recovered recently from Siberian soda lakes (Vavourakis et al. 2019). The final three members of the core community belonged to the class Clostridia, class Desulfuromonadia, and class Chrysiogenetes, which includes Peptostreptococcales-Tissierellales, Desulfuromusa, and Desulfurispirillum, respectively. The relative abundance of each core community member is shown in Fig. 2B. The most abundant bacterium was Thioalkalivibrio sulfidiphilus with a relative abundance of up to 60%.
To visualize the phylogenetic affiliation of the core community members, a phylogenetic tree was created with the 30 ASVs with their cultured and uncultured close relatives (Fig. 3). The tree shows that the members of the core community are affiliated with various phyla.
Prediction of the functional potential of the core community
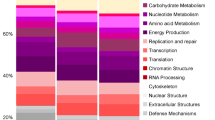
Using Tax4Fun2, we focused on the genes involved in the oxidation of reduced sulfur compounds. The predictions indicate the presence of all genes needed for complete sulfide oxidation to sulfur (Fig. 4; Table S2). The sulfide-oxidizing genes sqr, fccAB (Griesbeck et al. 2000; Trüper et al. 2002), and thiosulfate-oxidizing genes soxABXYZ were found to be highly abundant in the reactor samples from the paper mill (PM) and from the landfill (LF). We also searched for genes involved in the conversion of contaminants such as ammonia, thiols, and BTEX. Only BTEX degrading genes were found to be present in low numbers in oil and gas plants and pilots plant (Table S2).
Co-occurrence analysis
To understand the interaction among the members of the core community, a co-occurrence network was created. Figure 5 shows the co-occurrence of the core community members that have significant interactions. The network shows a negative correlation between the Thioalkalivibrio sulfidiphilus and Alkalilimnicola ehrlichii genera, a positive correlation between Alkalilimnicola ehrlichii and Halomonas sp. Likewise, Lentimicrobium, Acholeplasma, and Rhodobacteracea were found to have significant positive correlations in their occurrence. This relationship was also found for the Blvii-wastewater sludge group, Candidatus “Izimaplasma,” and members of the family Peptostreptococcales-Tissierellales.
Relation between physicochemical parameters and microbial community composition
Redundancy analysis was performed to find a relationship between the microbial community composition and the physicochemical parameters. The parameters that were significant explain the microbial composition with thiosulfate (R2 = 0.24, p < 0.05), sulfate (R2 = 0.30, p < 0.05) and alkalinity (R2 = 0.46, p < 0.05). As shown in Fig. 6, Guyparkeria, Nitrinicola, and Halomonas were associated with the presence of more sulfate and thiosulfate in the samples while the presence of Tv. halophilus and Alkalilimnicola were more aligned with alkalinity. On the other hand, Roseinatronbacter, Tv. sulfidiphilus, and Thiomicrospira were not associated with any of the tested parameters.
To find the differential abundance of the microbial population with respect to industry type, alkalinity, sulfate, and thiosulfate, the multinomial regression model was fitted with these variables. A fit value of Q2 = 0.636 was obtained. However, when the model was fitted with only alkalinity, sulfate, and thiosulfate, a fit value of Q2 = 0.09 was obtained suggesting that these factors alone cannot explain the differential abundances of microbial community members across the samples.
Discussion
Physicochemical analyses
In this paper, we have studied several operational and physicochemical parameters associated with BD reactors. Table 1 clearly indicates distinct variability of parameter values in all reactors and served as a good data set for studying the microbial composition across the plants. For the process, a higher rate of accumulation of sulfate and thiosulfate can be seen as an indicator of a decrease in sulfur selectivity (de Rink et al. 2019). For example, the measured levels of thiosulfate levels are indicators for dominating side-product oxidation pathways. Typically, thiosulfate formation cannot be avoided as, when dissolved oxygen and sulfide will chemically react to thiosulfate (de Graaff et al. 2012; Klok et al. 2013). Increased thiosulfate levels indicate that significant amounts of sulfide are chemically oxidized. Furthermore, the absence of thiosulfate indicates that all thiosulfate which is formed is subsequently oxidized to sulfate. While the analyzed concentrations of accumulated ions cannot directly be translated to the rate formation, these results can still be directly related to BD plant performance. Hence, these values are useful to explain the microbial community composition of the plants. For future analyses, the ratio of S2O32–/SO42– can also help in explaining the presence of SOB that are sulfide specialized and/or more generalists in the BD reactors. Higher S2O32–/SO42– reflects more chemical oxidation and can also suggest less thiosulfate oxidizing bacteria in the reactor. Additionally, the sampling and physicochemical analyses can be extended to have higher sampling frequencies to visualize the fluctuations in the reactors.
The ORP is considered to play an important role in determining sulfur selectivity. However, recently it has been shown that a constant redox potential does not lead to the highest sulfur selectivity, and a stoichiometry-driven feedforward controlling of oxygen has been suggested to lead to a high sulfur selectivity (Kiragosyan et al. 2020b). Nevertheless, microbial community composition can influence or can be influenced by the ORP and could indirectly affect the sulfur selectivity.
Microbial diversity analysis
The alpha diversity analyses have indicated that the two pilot plant samples (Pil1 and Pil2) have more diversity as compared to the full-scale BD reactors. This suggests that the addition of an anaerobic bioreactor might provide a niche to specific bacteria, for example, facultative anaerobes (de Rink et al. 2019) that were not present in the full-scale reactors.
The beta diversity analysis of the samples clearly suggests that the microbial communities are significantly affected by the composition and the source of gas of the different industries (Fig. 1). Having an input gas without hydrocarbon contaminants, the biomass of the paper mill (PM1, 2, 3, and 4) and landfill for building material which is more hydrocarbon free unlike usual landfills (LF1), these BD reactors were found to be very similar in their microbial composition while the biomass of the two oil and gas reactors (OG1 and 2) differs from each other in composition as they have distinct contaminants in them. Other studies have shown that the presence of thiols and BTEX have an effect on the microbial community composition in BD reactors (Roman et al., 2016; Kiragosyan et al. 2020a).
The microbial community was found to be less diverse in the BD-reactors that treat feed gas from the same source industry type suggesting the huge impact of the feed gas source. For example, Tv. sulfidiphilus was abundant in all paper mill and landfill plants (PM1–4, LF1), while Thiomicrospira was more abundant in pilot plant samples (Pil1 and 2). Other process parameters such as the sulfur separation method and the H2S/CO2 ratio have little effect on the microbial community (Supplementary Fig. S2). However, analysis with more BD reactors could help to get more conclusive results.
The core microbial community of BD reactors
The microbial diversity analysis of ten BD reactors clearly reveals a shared microbial community composition that is variably abundant across the plants. The 30 ASVs can be defined as the core microbial community because they are shared by all the plants and has no bias of their geographical location, the industrial gas streams they treat, or the differences in the physicochemical parameters. The possible reason for the presence of a core community is the haloalkaline environment in all the BD reactors. The microbiome of soda lakes, a natural haloalkaline environment, also has a core microbiome in lakes located across the continents (Zorz et al. 2019). A metagenome-based study of soda lakes has also highlighted the presence of similar phyla in the soda lakes that were found in BD reactors (Vavourakis et al. 2016). Interestingly, these 30 core community members are not only common but together also comprise a very large proportion of the total microbial diversity of BD reactors (Fig. 2B).
Taxonomy-based and phylogenetic analyses
Taxonomy-based assignment of the 30 core ASVs (Figs. 2B and 3) indicated that they could be assigned to the Gammaproteobacteria (16), Alphaproteobacteria (4), Bacteroidia (4), Bacilli (3), Clostridia (1), Desulfuromondia (1), and Chrysiogenetes (1). It is worth noting that within 30 core ASVs, there are ASVs that were assigned similar phylogenies even though they have different sequences. BLASTn analysis of these sequences has revealed that sequences vary in identity (96 to 100%) and coverage (90 to 100%). Based on this analysis, it is possible that they represent different species of bacteria or different strains of the same species. However, considering the fact that most common SOB such as T. sulfidiphilus and A. ehrlichii are not very phylogenetically distant (Ahn et al. 2017), we have considered all 30 ASVs as individual bacteria.
In general, several known SOBs belong to the class Gammaproteobacteria such as species of Thioalkalivibrio, Thiomicrospira, Alkalilimnicola, Guyparkeria, Thioalkalibacter, and Alkalispirillum (Rijkenberg et al. 2001; Sorokin et al. 2001, 2012; Hoeft et al. 2007; Banciu et al. 2008; Boden 2017). The first four were also part of the core community, indicative of their important role in sulfide oxidization in the BD process (Fig. 7), which was confirmed by the predicted abundance of the sqr and fccAB genes (Fig. 4). The later ones were only abundantly present in a few reactors. In addition, the core community also consisted of other members assigned as Ectothiorhodospira. Members of this phototrophic genus can oxidize sulfur compounds. Other genera belonging to a group of 16 ASVs contained Halomonas, Nitrincola, and Wenzhouxiangella. Halomonas are known organotrophs but can also oxidize sulfur compounds incompletely to tetrathionate (Sorokin 2003). Some of the denitrifying species have the potential to oxidize sulfide and thiosulfate anaerobically (Sorokin 2003; Wang and Shao 2021). Nitrincola is a facultatively anaerobic, chemo-organotrophic bacterium capable of nitrate reduction but not known for sulfide oxidation. But they contain potential genes for thiosulfate dehydrogenase yielding tetrathionate (Vavourakis et al. 2016; Borsodi et al. 2017). A novel haloalkaliphilic species of the genus Wenzhouxiangella sp. has recently been enriched and isolated from Siberian soda lakes using cells of Gram-positive cocci as substrate and was proven to be strongly proteolytic with the ability to feed on microbial biomass (Sorokin et al. 2020). In all, these obligate heterotrophic haloalkaliphiles can be considered a “satellite” population of the autotrophic SOB feeding on their organic products with an ability to marginally contribute to sulfur cycling, such as by the production of tetrathionate.
Members of the class Alphaproteobacteria and family Rhodobacteraceae such as Roseinatronbacter are aerobic sulfur-oxidizing lithoheterotrophs that oxidize inorganic sulfur compounds to sulfate during organotrophic growth (Sorokin et al. 2000; Boldareva et al. 2007; Gorlenko et al. 2010). Stappia is well known to be involved in denitrification and CO oxidation (Weber and King 2007). However, it has genes to convert thiosulfate to sulfate (Huang et al. 2015a) and has been found in several sulfide-removing reactors (Huang et al. 2015b; San-Valero et al. 2019). Their role in BD reactors is shown in Fig. 7.
The ASVs assigned to Bacteroidota include Planktosalinus, Lentimicobium, uncultured_Bacteroidetes/Chlorobi, and Psychroflexus. So far only one member of Planktosalinus, P. lacus has been characterized as a strict aerobic heterotroph (Zhong et al. 2016). For Lentimicrobium, L. saccharophilum is a strict anaerobe and cannot use any sulfur compounds as electron acceptors (Sun et al. 2016). Uncultured_Bacteroidetes/Chlorobi assigned to the Blvii28_wastewater-sludge_group are members of the core community, and they are frequently detected in sulfide-oxidizing bioreactors (Vannini et al. 2008). However, the specific role of this Blvii28_wastewater-sludge_group is not clear. On the other hand, Chlorobi, the green sulfur bacteria, are known to perform sulfide oxidation (Fig. 7) (Visser et al. 1997; Ghosh and Dam 2009). Members of the genus Psychroflexus and, in particular, P. torquis are very versatile bacteria that use an extensive range of carbon and energy sources, including carbohydrates, amino acids, organic acids, and odd chain length lipid oxidation products (Bowman et al. 1998).
Members of class Bacilli namely Acholeplasma and Candidatus “Izimaplasma” are not known to participate in the sulfur cycle but may play a role in degrading organic matter present in the BD reactors (Skennerton et al. 2016; de Rink et al. 2019; Zorz et al. 2019). Member of class Clostridia, order Peptostreptococcales-Tissierellales are also part of the core community. Some of these members are strictly anaerobic and can reduce elemental sulfur (Fig. 7) (Takai et al. 2001), and some are haloalkaliphilic acetogens that cannot reduce sulfate (Pikuta et al. 2003). Lastly, members of the class Desulfuromondia and Chrysiogenetes, namely, Desulfuromosa and Desulfurisprillum are anaerobic organotrophs and can use elemental sulfur as an electron acceptor (Fig. 7) (Liesack and Finster 1994; Sorokin et al. 2007).
Figure 7 summarizes the possible biological conversions that involve one or more core members in the BD reactors, and Table 2 lists the bacteria associated with these conversions. The figure suggests that the core community consisted of bacteria with diverse functions. While for some SOB, the role can be clearly associated with several oxidation steps of sulfide, there are also thiosulfate oxidizers and sulfur reducers present but no sulfate reducers. There are also several members which are heterotrophs and whose potential role in sulfur conversions is not known; however, metagenomic analyses of natural and other sulfide-related reactors have also listed some of these bacteria. This could mean that these bacteria use the organic matter from decomposed bacterial cells and their role in the reactor is an opportunist scavenger. Tetrathionate could also be formed by heterotrophic bacteria from thiosulfate oxidation (Sorokin 2003). Tetrathionate is instable at high pH (>10) and is chemically decomposed to thiosulfate, trithionate, and pentathionate (Varga and Horváth 2007). It can also react with sulfide to form sulfur and thiosulfate. Furthermore, some bacteria can biologically convert tetrathionate to sulfur, sulfate, and thiosulfate (Sorokin 2003). Due to the above-mentioned conversions, tetrathionate is less likely to be available as an intermediate in the BD reactors and therefore contributes little, if at all, to the sulfur conversions. This is also shown by the low abundance of the genes involved in tetrathionate conversions (Table S2).
In addition to the biological reactions, Figure 7 also shows the possible chemical conversions that play an important role in generating alternative substrates for SOB, like polysulfides and thiosulfate.
The core ASVs shape the major fraction of the total microbial community, but there are other species that were abundant only in some of the BD reactors (Fig. 2B). Alkalisprillum was abundant (14.3%) in Papermill_02, Vibrio (16.2%) in Papermill_03 and Nitrincola (13.8%) in Cadaver_01. Alkalisprillum is a close relative of Alkalilimnicola and is known to oxidize sulfide/polysulfide to elemental sulfur aerobically (Sorokin et al. 2006). Members of genera Vibrio are widely found in marine and haloalkaline environments (Simidu and Tsukamoto 1985; Mwirichia et al. 2010). Studies have predicted that members of the family Vibrionacea have the potential to oxidize thiosulfate to tetrathionate with heterotrophic growth (Sorokin 2003).
Prediction of the functional potential of the core community
Tax4Fun2 predicted the overall high abundance of genes involved in the oxidation of sulfide to sulfur (sqr; fccAB; Fig. 4). However, sqr and fccAB were variably present in different reactors suggesting that the mechanism of sulfide oxidation might differ among the different industry reactors. The possibility of variable mechanisms was also shown in the previous studies on the sludge from full-scale plants (Kiragosyan et al. 2019). Additionally, paper mill (PM1–4) and landfill-treating reactors (LF1) have a similar functional profile. These plants were dominated by Thioalkalivibrio sulfidiphilus which has fccAB genes and a sox gene system but lacks the sqr gene. Contrarily, the rendering waste treating reactor (Cad1) has a very different functional profile than others with a high abundance of sqr genes and less of fcc and sox genes. The pilot runs studied in this research have an abundance of both fcc and sqr. These results differ from the previous research where sqr was suggested to be most abundant (de Rink et al. 2019). All these predictions indicate the differences in the potential mechanisms of sulfide oxidation among the reactors. However, to make a definite conclusion, more information on the sulfide loads and sulfur selectivity along with gene expression results is needed.
Co-occurrence analyses
Co-occurrence analysis of the core community has revealed an antagonistic relationship between Tv. sulfidiphilus and A. ehrlichii (Fig. 5). Recent studies with the modified BD line-up have also indicated these dynamics in the reactors (de Rink et al. 2019). Being both a facultative autotroph and facultative anaerobe, A. ehrlichii is more versatile than T. sulfidiphilus, which is a strict autotroph. A. ehrlichii was found to co-occur with Halomonas sp., indicating the presence of organic compounds in the reactor. The analysis also shows that there are two sets of heterotrophic bacteria from different phyla co-occurring in the BD reactors.
Relation between physicochemical parameters and microbial community composition
Though the biological process of full-scale BD reactors is the same, there are several operational and design parameters that make them different. RDA analysis indicated that the concentration of thiosulfate, sulfate, and alkalinity in the BD reactors is related to the presence of certain bacteria. Figure 7 highlights that members of the core community have a different potential to oxidize sulfur compounds. The complete/incomplete biological sulfide oxidation can thus lead to the formation of sulfate/sulfur. Moreover, different SOBs, such as Thioalkalivibrio and Thiomicrospira have different growth strategies (Sorokin et al. 2003).
Biological and chemical conversions occurring in the BD reactors. The numbers represent the biological conversions performed by the core community members. The letters denote the chemical reactions. The description of the numbers and letters are specified in Table 2
From the results of RDA analysis (Fig. 6), it is evident that Guyparkeria and Halomonas were found to be highly correlated with the increasing concentration of thiosulfate and sulfate in the BD process. Guyparkeria can oxidize several reduced sulfur compounds and oxidize them to sulfate (Sievert et al. 2000) and can decrease the alkalinity of a neutral environment They are also capable to drive a neutral environment to less alkalinity (Whaley-Martin et al. 2019). This can be explained by an increase in H+ ions concurrently formed with sulfate production. On the other hand, Alkalilimnicola was more correlated with high alkalinity.
As mentioned before, in a BD reactor a high rate of thiosulfate and sulfate formation together with their accumulated concentration indicates a sub-optimal process performance. Nevertheless, considering only the accumulated concentration, Guyparkeria and Halomonas can be regarded as indicator organisms of high sulfate and thiosulfate accumulation in the process. High thiosulfate and sulfate formation directly corresponds to a decreased alkalinity and concomitantly an increase in the caustic demand, thereby decreasing the performance of the BD process and increasing costs.
To conclude, microbial community analysis of ten full-scale BD reactors treating gas streams from different industries demonstrated the presence of a core microbiome consisting of 30 ASVs with Thioalkalivibrio sulfidiphilus as the most abundant community member. The diversity is largely affected by the source and composition of the feed gas. Members of the genera Guyperkaria and Halomonas are related to an increase in sulfate and thiosulfate levels and as such can be regarded as indicator organisms for a suboptimal performance of the BD processes.
In this study, only a few physicochemical parameters such as pH, salinity, alkalinity, conductivity, thiosulfate, sulfate concentrations have been considered. In the future, it is recommended to include parameters such as ORP, sulfide load, airflow rate, and sulfur selectivity while performing microbial composition analysis for these reactors.
Availability of data and material
All the data and material has been shared as part of the manuscript. The amplicon sequences of this study can be found in the Sequence Read Archive under PRJEB43104.
Code availability
Not applicable.
References
Ahn AC, Meier-Kolthoff JP, Overmars L, Richter M, Woyke T, Sorokin DY, Muyzer G (2017) Genomic diversity within the haloalkaliphilic genus Thioalkalivibrio. PLoS ONE 12:1–23. https://doi.org/10.1371/journal.pone.0173517
Banciu HL, Sorokin DY, Tourova TP, Galinski EA, Muntyan MS, Kuenen JG, Muyzer G (2008) Influence of salts and pH on growth and activity of a novel facultatively alkaliphilic, extremely salt-tolerant, obligately chemolithoautotrophic sufur-oxidizing Gammaproteobacterium Thioalkalibacter halophilus gen. nov., sp. nov. from South-Western Siber. Extremophiles 12:391–404. https://doi.org/10.1007/s00792-008-0142-1
Berben T, Overmars L, Sorokin DY, Muyzer G (2019) Diversity and distribution of sulfur oxidation-related genes in Thioalkalivibrio, a genus of chemolithoautotrophic and haloalkaliphilic sulfur-oxidizing bacteria. Front Microbiol 10:160. https://doi.org/10.3389/fmicb.2019.00160
Boden R (2017) Reclassification of Halothiobacillus hydrothermalis and Halothiobacillus halophilus to Guyparkeria gen. nov. in the Thioalkalibacteraceae fam. nov., with emended descriptions of the genus Halothiobacillus and family Halothiobacillaceae. Int J Syst Evol Microbiol 67:3919–3928. https://doi.org/10.1099/ijsem.0.002222
Boden R, Scott KM, Williams J, Russel S, Antonen K, Rae AW, Hutt LP (2017) An evaluation of Thiomicrospira, Hydrogenovibrio and Thioalkalimicrobium: reclassification of four species of Thiomicrospira to each Thiomicrorhabdus gen. nov. and Hydrogenovibrio, and reclassification of all four species of Thioalkalimicrobium to Thiomicrospira. Int J Syst Evol Microbiol 67:1140–1151. https://doi.org/10.1099/ijsem.0.001855
Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:1–17. https://doi.org/10.1186/s40168-018-0470-z
Boldareva EN, Bryantseva IA, Tsapin A, Nelson K, Sorokin DY, Tourova TP, Boichenko VA, Stadnichuk IN, Gorlenko VM (2007) The new alkaliphilic bacteriochlorophyll a-containing bacterium Roseinatronobacter monicus sp. nov. from the hypersaline soda Mono Lake (California, United States). Microbiology 76:82–92. https://doi.org/10.1134/S0026261707010122
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Bin KK, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Borsodi AK, Korponai K, Schumann P, Spröer C, Felföldi T, Márialigeti K, Szili-Kovács T, Tóth E (2017) Nitrincola alkalilacustris sp. nov. and Nitrincola schmidtii sp. nov., alkaliphilic bacteria isolated from soda pans, and emended description of the genus Nitrincola. Int J Syst Evol Microbiol 67:5159–5164. https://doi.org/10.1099/ijsem.0.002437
Bowman JP, Mccammon SA, Lewis T, Skerratt JH, Brown JL, Nichols DS, Mcmeekin TA (1998) Psychrophilic species from Antarctic sea ice and reclassification of Flavobacterium. Microbiology 144:1601–1609
Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, A AJ (2016) Dada2. 13:581–583. https://doi.org/10.1038/nmeth.3869.DADA2
de Graaff M, Bijmans MFM, Abbas B, Euverink GJW, Muyzer G, Janssen AJH (2011) Biological treatment of refinery spent caustics under halo-alkaline conditions. Bioresour Technol 102:7257–7264. https://doi.org/10.1016/j.biortech.2011.04.095
de Graaff M, Klok JBM, Bijmans MFM, Muyzer G, Janssen AJH (2012) Application of a 2-step process for the biological treatment of sulfidic spent caustics. Water Res 46:723–730. https://doi.org/10.1016/j.watres.2011.11.044
de Rink R, Klok JBM, van Heeringen GJ, Keesman KJ, Janssen AJH, ter Heijne A, Buisman CJN (2020) Biologically enhanced hydrogen sulfide absorption from sour gas under haloalkaline conditions. J Hazard Mater 383:121104. https://doi.org/10.1016/j.jhazmat.2019.121104
de Rink R, Klok JBM, van Heeringen GJ, Sorokin DY, ter Heijne A, Zeijlmaker R, Mos YM, de Wilde V, Keesman KJ, Buisman CJN (2019) Increasing the selectivity for sulfur formation in biological gas desulfurization. Environ Sci Technol 53:4519–4527. https://doi.org/10.1021/acs.est.8b06749
Ghosh W, Dam B (2009) Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol Rev 33:999–1043. https://doi.org/10.1111/j.1574-6976.2009.00187.x
Gorlenko VM, Buryukhaev SP, Matyugina EB, Borzenko SV, Namsaraev ZB, Bryantseva IA, Boldareva EN, Sorokin DY, Namsaraev BB (2010) Microbial communities of the stratified soda Lake Doroninskoe (Transbaikal region). Microbiology 79:390–401. https://doi.org/10.1134/S0026261710030161
Griesbeck C, Hauska G, Schutz M (2000) Biological sulfide oxidation: sulfide-quinone-reductase (SQR), the Primary Reaction, Vol. 4, Research Signpost, Trivandrum, India.
Heberle H, Meirelles VG, da Silva FR, Telles GP, Minghim R (2015) InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16:1–7. https://doi.org/10.1186/s12859-015-0611-3
Hoeft SE, Blum JS, Stolz JF, Tabita FR, Witte B, King GM, Santini JM, Oremland RS (2007) Alkalilimnicola ehrlichii sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammaproteobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int J Syst Evol Microbiol 57:504–512. https://doi.org/10.1099/ijs.0.64576-0
Huang C, Li Z, Chen F, Liu Q, Zhao Y, Zhou J, Wang A (2015a) Microbial community structure and function in response to the shift of sulfide/nitrate loading ratio during the denitrifying sulfide removal process. Bioresour Technol 197:227–234. https://doi.org/10.1016/j.biortech.2015.08.019
Huang C, Zhao Y, Li Z, Yuan Y, Chen C, Tan W, Gao S, Gao L, Zhou J, Wang A (2015b) Enhanced elementary sulfur recovery with sequential sulfate-reducing, denitrifying sulfide-oxidizing processes in a cylindrical-type anaerobic baffled reactor. Bioresour Technol 192:478–485. https://doi.org/10.1016/j.biortech.2015.04.103
Janssen AJH, Lens PNL, Stams AJM, Plugge CM, Sorokin DY, Muyzer G, Dijkman H, van Zessen E, Luimes P, Buisman CJN (2009) Application of bacteria involved in the biological sulfur cycle for paper mill effluent purification. Sci Total Environ 407:1333–1343. https://doi.org/10.1016/j.scitotenv.2008.09.054
Janssen AJH, Ruitenberg R, Buisman CJN (2001) Industrial applications of new sulphur biotechnology. Water Sci Technol 44:85–90. https://doi.org/10.2166/wst.2001.0471
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kiragosyan K, Klok JBM, Keesman KJ, Roman P, Janssen AJH (2019) Development and validation of a physiologically based kinetic model for starting up and operation of the biological gas desulfurization process under haloalkaline conditions. Water Res X 4:100035. https://doi.org/10.1016/j.wroa.2019.100035
Kiragosyan K, Picard M, Timmers PHA, Sorokin DY, Klok JBM, Roman P, Janssen AJH (2020a) Effect of methanethiol on process performance, selectivity and diversity of sulfur-oxidizing bacteria in a dual bioreactor gas biodesulfurization system. J Hazard Mater 398:123002. https://doi.org/10.1016/j.jhazmat.2020.123002
Kiragosyan K, Roman P, Keesman KJ, Janssen AJH, Klok JBM (2020b) Stoichiometry-driven heuristic feedforward control for oxygen supply in a biological gas desulfurization process. J Process Control 94:36–45. https://doi.org/10.1016/j.jprocont.2020.08.007
Klok JBM, van Heeringen GJ, de Rink R, Wijnbelt J (2017) Introducing the next generation of the THIOPAQ R&G process for biotechnological gas desulphurization: THIOPAQ-SQ. Sulfur 1–4
Klok JBM, de Graaff M, van den Bosch PLF, Boelee NC, Keesman KJ, Janssen AJH (2013) A physiologically based kinetic model for bacterial sulfide oxidation. Water Res 47:483–492. https://doi.org/10.1016/j.watres.2012.09.021
Klok JBM, van Heeringen GJ, de Rink R, Wijnbelt H, Bowerbank G (2018) Techno-economic impact of the next generation Thiopaq O&G process for sulfur removal. In: Proceedings of the GPA-GCC 26th Annual Technical Conference. Muscat
Liesack W, Finster K (1994) Phylogenetic analysis of five strains of Gram-negative, obligately anaerobic, sulfur-reducing bacteria and description of Desulfuromusa gen. nov., including Desulfuromusa kysingii sp. nov., Desulfuromusa bakii sp. nov., and Desulfuromusa succinoxidans sp. Int J Syst Bacteriol 44:753–758. https://doi.org/10.1099/00207713-44-4-753
Mahmood Q, Zheng P, Cai J, Hayat Y, Hassan M, Wu D, Hu B (2007) Sources of sulfide in waste streams and current biotechnologies for its removal. J Zhejiang Univ - Sci A 8:1126–1140. https://doi.org/10.1631/jzus.2007.A1126
Mol AR, van der Weijden RD, Klok JBM, Buisman CJN (2020) Properties of sulfur particles formed in biodesulfurization of biogas. Minerals 10: https://doi.org/10.3390/min10050433
Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R (2019) Establishing microbial composition measurement standards with reference frames. Nat Commun 10. https://doi.org/10.1038/s41467-019-10656-5
Muyzer G, Kuenen JG, Robertson LA (2013) Colorless sulfur bacteria. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes: Prokaryotic physiology and biochemistry. Springer, Berlin Heidelberg, Berlin, Heidelberg, pp 555–588
Muyzer G, Stams AJM (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 45:187–192. https://doi.org/10.1038/nrmicro1892
Mwirichia R, Muigai AW, Tindall B, Boga HI, Stackebrandt E (2010) Isolation and characterisation of bacteria from the haloalkaline Lake Elmenteita, Kenya. Extremophiles 14:339–348. https://doi.org/10.1007/s00792-010-0311-x
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Olivier G, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A (2011) Machine learning in Python. J Mach Learn Res 12:2825–2830
Pikuta EV, Hoover RB, Bej AK, Marsic D, Detkova EN, Whitman WB, Krader P (2003) Tindallia californiensis sp. nov., a new anaerobic, haloalkaliphilic, spore-forming acetogen isolated from Mono Lake in California. Extremophiles 7:327–334. https://doi.org/10.1007/s00792-003-0326-7
Po HN, Senozan NM (2001) The Henderson-Hasselbalch equation: its history and limitations. J Chem Educ 78:1499–1503. https://doi.org/10.1021/ed078p1499
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS ONE 5. https://doi.org/10.1371/journal.pone.0009490
Pruesse E, Peplies J, Glöckner FO (2012) SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. https://doi.org/10.1093/bioinformatics/bts252
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. https://doi.org/10.1093/nar/gks1219
Rijkenberg MJA, Kort R, Hellingwerf KJ (2001) Alkalispirillum mobile gen. nov., spec. nov., an alkaliphilic non-phototrophic member of the Ectothiorhodospiraceae. Arch Microbiol 175:369–375. https://doi.org/10.1007/s002030100274
Roman P, Klok JBM, Sousa JAB, Broman E, Dopson M, van Zessen E, Bijmans MFM, Sorokin DY, Janssen AJH (2016) Selection and application of sulfide oxidizing microorganisms able to withstand thiols in gas biodesulfurization systems. Environ Sci Technol 50:12808–12815. https://doi.org/10.1021/acs.est.6b04222
Roman P, Veltman R, Bijmans MFM, Keesman KJ, Janssen AJH (2015) Effect of methanethiol concentration on sulfur production in biological desulfurization systems under haloalkaline conditions. Environ Sci Technol 49:9212–9221. https://doi.org/10.1021/acs.est.5b01758
San-Valero P, Penya-roja JM, Javier Álvarez-Hornos F, Buitrón G, Gabaldón C, Quijano G (2019) Fully aerobic bioscrubber for the desulfurization of H2S-rich biogas. Fuel 241:884–891. https://doi.org/10.1016/j.fuel.2018.12.098
Sievert SM, Heidorn T, Kuever J (2000) Halothiobacillus kellyi sp. nov., a mesophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium isolated from a shallow- water hydrothermal vent in the Aegean Sea, and emended description of the genus Halothiobacillus. Int J Syst Evol Microbiol 50:1229–1237. https://doi.org/10.1099/00207713-50-3-1229
Simidu U, Tsukamoto K (1985) Habitat segregation and biochemical activities of marine members of the family Vibrionaceae. Appl Environ Microbiol 50:781–790. https://doi.org/10.1128/aem.50.4.781-790.1985
Skennerton CT, Haroon MF, Briegel A, Shi J, Jensen GJ, Tyson GW, Orphan VJ (2016) Phylogenomic analysis of Candidatus “Izimaplasma” species: free-living representatives from a Tenericutes clade found in methane seeps. ISME J 10:2679–2692. https://doi.org/10.1038/ismej.2016.55
Sorokin DY (2003) Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72:641–653. https://doi.org/10.1023/B:MICI.0000008363.24128.e5
Sorokin DY, Banciu H, Robertson LA, Kuenen JG, Muntyan MS, Muyzer G (2013) Halophilic and haloalkaliphilic sulfur-oxidizing bacteria. In: The Prokaryotes: Prokaryotic Physiology and Biochemistry. pp 529–554
Sorokin DY (2002) Thioalkalispira microaerophila gen. nov., sp. nov., a novel lithoautotrophic, sulfur-oxidizing bacterium from a soda lake. Int J Syst Evol Microbiol 52:2175–2182. https://doi.org/10.1099/ijs.0.02339-0
Sorokin DY, Banciu H, van Loosdrecht M, Kuenen JG (2003) Growth physiology and competitive interaction of obligately chemolithoautotrophic, haloalkaliphilic, sulfur-oxidizing bacteria from soda lakes. Extremophiles 7:195–203. https://doi.org/10.1007/s00792-002-0313-4
Sorokin DY, Banciu HL, Muyzer G (2015) Functional microbiology of soda lakes. Curr Opin Microbiol 25:88–96. https://doi.org/10.1016/j.mib.2015.05.004
Sorokin DY, Foti M, Tindall BJ, Muyzer G (2007) Desulfurispirillum alkaliphilum gen. nov. sp. nov., a novel obligately anaerobic sulfur- and dissimilatory nitrate-reducing bacterium from a full-scale sulfide-removing bioreactor. Extremophiles 11:363–370. https://doi.org/10.1007/s00792-006-0048-8
Sorokin DY, Lysenko AM, Mityushina LL, Tourova TP, Jones BE, Rainey FA, Robertson LA, Kuenen GJ (2001) Thioalkalimicrobium aerophilum gen. nov., sp. nov. and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp. nov. and Thioalkalivibrio denitrificans sp. nov., novel obligately alkaliphilic. Int J Syst Evol Microbiol 51:565–580. https://doi.org/10.1099/00207713-51-2-565
Sorokin DY, Mosier D, Zorz JK, Dong X, Strous M (2020) Wenzhouxiangella strain AB-CW3, a proteolytic bacterium from hypersaline soda lakes that preys on cells of gram-positive bacteria. Front Microbiol 11:1–14. https://doi.org/10.3389/fmicb.2020.597686
Sorokin DY, Muntyan MS, Panteleeva AN, Muyzer G (2012) Thioalkalivibrio sulfidiphilus sp. nov., a haloalkaliphilic, sulfur-oxidizing gammaproteobacterium from alkaline habitats. Int J Syst Evol Microbiol 62:1884–1889. https://doi.org/10.1099/ijs.0.034504-0
Sorokin DY, Tourova TP, Kuznetsov BB, Bryantseva IA, Gorlenko VM (2000) Roseinatronobacter thiooxidans gen. nov., sp. nov., a new alkaliphilic aerobic bacteriochlorophyll a-containing bacterium isolated from a soda Lake. Microbiology 69:89–97. https://doi.org/10.1007/BF02757261
Sorokin DY, van den Bosch PLF, Abbas B, Janssen AJH, Muyzer G (2008) Microbiological analysis of the population of extremely haloalkaliphilic sulfur-oxidizing bacteria dominating in lab-scale sulfide-removing bioreactors. Appl Microbiol Biotechnol 80:965–975. https://doi.org/10.1007/s00253-008-1598-8
Sorokin DY, Zhilina TN, Lysenko AM, Tourova TP, Spiridonova EM (2006) Metabolic versatility of haloalkaliphilic bacteria from soda lakes belonging to the Alkalispirillum-Alkalilimnicola group. Extremophiles 10:213–220. https://doi.org/10.1007/s00792-005-0487-7
Sun L, Toyonaga M, Ohashi A, Tourlousse DM, Matsuura N, Meng XY, Tamaki H, Hanada S, Cruz R, Yamaguchi T, Sekiguchi Y (2016) Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum bacteroidetes, and proposal of Lentimicrobiaceae fam. nov. Int J Syst Evol Microbiol 66:2635–2642. https://doi.org/10.1099/ijsem.0.001103
Takai K, Moser DP, Onstott TC, Spoelstra N, Pfiffner SM, Dohnalkova A, Fredrickson JK (2001) Alkaliphilus transvaalensis gen. nov., sp. nov., an extremely alkaliphilic bacterium isolated from a deep South African gold mine. Int J Syst Evol Microbiol 51:1245–1256. https://doi.org/10.1099/00207713-51-4-1245
Tong WM, Chan Y (2020) GenePiper, a graphical user interface tool for microbiome sequence data mining. Microbiol Resour Announc 9:13–15. https://doi.org/10.1128/mra.01195-19
Trüper HG, Brüser T, Tschäpe J, Reinartz M, Dahl C (2002) Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol 170:59–68. https://doi.org/10.1007/s002030050615
van den Bosch PLF, van Beusekom OC, Buisman CJN, Janssen AJH (2007) Sulfide oxidation at halo-alkaline conditions in a fed-batch bioreactor. Biotechnol Bioeng 97:1053–1063. https://doi.org/10.1002/bit.21326
Vannini C, Munz G, Mori G, Lubello C, Verni F, Petroni G (2008) Sulphide oxidation to elemental sulphur in a membrane bioreactor: performance and characterization of the selected microbial sulphur-oxidizing community. Syst Appl Microbiol 31:461–473. https://doi.org/10.1016/j.syapm.2008.07.003
Varga D, Horváth AK (2007) Kinetics and mechanism of the decomposition of tetrathionate ion in alkaline medium. Inorg Chem 46:7654–7661. https://doi.org/10.1021/ic700992u
Vavourakis CD, Ghai R, Rodriguez-Valera F, Sorokin DY, Tringe SG, Hugenholtz P, Muyzer G (2016) Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline soda lake brines. Front Microbiol 7:1–18. https://doi.org/10.3389/fmicb.2016.00211
Vavourakis CD, Mehrshad M, Balkema C, van Hall R, Andrei A-Ş, Ghai R, Sorokin DY, Muyzer G (2019) Metagenomes and metatranscriptomes shed new light on the microbial-mediated sulfur cycle in a Siberian soda lake. BMC Biol 17:1–20. https://doi.org/10.1186/s12915-019-0688-7
Visser JM, De Jong GAH, Robertson LA, Kuenen JG (1997) A novel membrane-bound flavocytochrome c sulfide dehydrogenase from the colourless sulfur bacterium Thiobacillus sp. W5. Arch Microbiol 167:295–301. https://doi.org/10.1007/s002030050447
Wang L, Shao Z (2021) Aerobic denitrification and heterotrophic sulfur oxidation in the genus Halomonas revealed by six novel species characterizations and genome-based analysis. Front Microbiol 12. https://doi.org/10.3389/fmicb.2021.652766
Weber CF, King GM (2007) Physiological, ecological, and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus. Appl Environ Microbiol 73:1266–1276. https://doi.org/10.1128/AEM.01724-06
Wemheuer F, Taylor JA, Daniel R, Johnston E, Meinicke P, Thomas T, Wemheuer B (2018) Tax4Fun2: a R-based tool for the rapid prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene marker gene sequences. bioRxiv 7:490037. https://doi.org/10.1101/490037
Whaley-Martin K, Jessen GL, Nelson TC, Mori JF, Apte S, Jarolimek C, Warren LA (2019) The potential role of Halothiobacillus spp. in sulfur oxidation and acid generation in circum-neutral mine tailings reservoirs. Front Microbiol 10. https://doi.org/10.3389/fmicb.2019.00297
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO (2014) The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:643–648. https://doi.org/10.1093/nar/gkt1209
Zhong ZP, Liu Y, Wang F, Zhou YG, Liu HC, Liu Q, Liu ZP (2016) Planktosalinus lacus gen. Nov., sp. nov., a member of the family flavobacteriaceae isolated from a salt lake. Int J Syst Evol Microbiol 66:2084–2089. https://doi.org/10.1099/ijsem.0.000997
Zorz JK, Sharp C, Kleiner M, Gordon PMK, Pon RT, Dong X, Strous M (2019) A shared core microbiome in soda lakes separated by large distances. Nat Commun 10:4230. https://doi.org/10.1038/s41467-019-12195-5
Acknowledgements
This work was performed in the cooperation framework of Wetsus, the European Centre of Excellence for Sustainable Water Technology (www.wetsus.nl). We thank all the full-scale biodesulfurization plant operators and Rieks de Rink for providing sludge for this study. We thank dr. Dimitri Y. Sorokin for critical discussions. We thank Pieter van Veelen for his help with the data analysis in QIIME 2 and R.
Funding
This work was performed in the cooperation framework of Wetsus, the European Centre of Excellence for Sustainable Water Technology (www.wetsus.nl). Wetsus is co-funded by the Dutch Ministry of Economic Affairs and Ministry of Infrastructure and Environment, the European Union Regional Development Fund, the Province of Fryslan, and the Northern Netherlands Provinces. This work is part of a project that has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skfodowska-Curie grant agreement no. 65874. The study was cofinanced by Paqell B.V.
Author information
Authors and Affiliations
Contributions
SG, CP, and GM conceived and designed the research. SG conducted the experiments. JK contributed in materializing the study by procuring samples and analyzing physicochemical data. SG and GM analyzed the data. SG wrote the manuscript. All authors contributed to the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gupta, S., Plugge, C.M., Klok, J.B.M. et al. Comparative analysis of microbial communities from different full-scale haloalkaline biodesulfurization systems. Appl Microbiol Biotechnol 106, 1759–1776 (2022). https://doi.org/10.1007/s00253-022-11771-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11771-y