Abstract
In this research, we were interested in answering a question whether subjecting a Yarrowia lipolytica strain overproducing a recombinant secretory protein (rs-Prot) to pre-optimized stress factors may enhance synthesis of the rs-Prot. Increased osmolarity (3 Osm kg−1) was the primary stress factor implemented alone or in combination with decreased temperature (20 °C), known to promote synthesis of rs-Prots. The treatments were executed in batch bioreactor cultures, and the cellular response was studied in terms of culture progression, gene expression and global proteomics, to get insight into molecular bases underlying an awaken reaction. Primarily, we observed that hyperosmolarity executed by high sorbitol concentration does not enhance synthesis of the rs-Prot but increases its transcription. Expectedly, hyperosmolarity induced synthesis of polyols at the expense of citric acid synthesis and growth, which was severely limited. A number of stress-related proteins were upregulated, including heat-shock proteins (HSPs) and aldo–keto reductases, as observed at transcriptomics and proteomics levels. Concerted downregulation of central carbon metabolism, including glycolysis, tricarboxylic acid cycle and fatty acid synthesis, highlighted redirection of carbon fluxes. Elevated abundance of HSPs and osmolytes did not outbalance the severe limitation of protein synthesis, marked by orchestrated downregulation of translation (elongation factors, several aa-tRNA synthetases), amino acid biosynthesis and ribosome biogenesis in response to the hyperosmolarity. Altogether we settled that increased osmolarity is not beneficial for rs-Prots synthesis in Y. lipolytica, even though some elements of the response could assist this process. Insight into global changes in the yeast proteome under the treatments is provided.
Key points
• Temp enhances, but Osm decreases rs-Prots synthesis by Y. lipolytica.
• Enhanced abundance of HSPs and osmolytes is overweighted by limited translation.
• Global proteome under Osm, Temp and Osm Temp treatments was studied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental factors evoke multilayered responses in the yeast producer cells. Some treatment conditions are known to promote synthesis of specific compounds, while the others exert adverse impact in this regard. For example, it is widely recognized that hyperosmotic conditions promote synthesis of polyols in yeast and that decreased temperature is beneficial for synthesis and secretion of recombinant, secretory proteins (rs-Prots). Both these phenomena were also observed and employed in bioprocesses with a nonconventional yeast species Yarrowia lipolytica (Tomaszewska et al. 2012; Yang et al. 2014, 2015; Kubiak et al. 2021), which is a popular workhorse in numerous industrial applications (Groenewald et al. 2014; Madzak 2018, 2021). Exploitation of Y. lipolytica as a rs-Prot expression platform receives significant attention due to several advantageous qualities of this species predisposing it to high level production (Celińska and Nicaud 2019; Theron et al. 2020). Up to date, several strategies have been adopted to further enhance synthesis of rs-Prots in this species, including invention of potent cloning tools (Schwartz et al. 2016; Celińska et al. 2017b, 2018; Trassaert et al. 2017; Wong et al. 2017; Park et al. 2019), different bioprocessing and medium optimization approaches (Chang et al. 1998a,b; Gasmi et al. 2011a,b; Celińska et al. 2017a), to a recent co-expression of omics-targeted secretory helpers that assist synthesis and secretion of rs-Prots (Korpys-Woźniak et al. 2021; Korpys-Woźniak and Celińska 2021). In our current studies, we are interested in answering a question whether subjecting a Y. lipolytica production culture to a specific, pre-optimized stress factor may enhance synthesis of rs-Prots. While the fact of beneficial impact of decreased temperature on synthesis and secretion of rs-Prots by yeast is widely recognized and largely generalizable, the impact of elevated osmolarity on the production has not been systematically addressed. Previous evidence suggests that increased osmotic pressure applied to cultures of filamentous fungi triggered enhanced synthesis of native proteins (Fiedurek 1998). Likewise, exposure of bacteria to high salt, sorbitol (SORB) and betaine concentrations increased production of a protein to be crystallized (Oganesyan et al. 2007). The outcome was attributed to increased cellular levels of osmolytes and chaperones, exerting protective activity toward polypeptides. Similar suggestions stemmed from a work on a recombinant Y. lipolytica strain expressing a heterologous invertase gene (Lazar et al. 2011). While the relationship between osmotic stress and enhanced synthesis of polyols in Y. lipolytica is clearly understood and well-described at molecular level (Kobayashi et al. 2015; Rzechonek et al. 2018), the effect of the former on the production of rs-Prots requires further studies.
In our previous experiments, we tested a working hypothesis on the beneficial effect of elevated osmotic pressure on the production of rs-Prots in Y. lipolytica (Kubiak et al. 2019). The hyperosmotic conditions were implemented by the addition of several chemically different osmoactive compounds, including glycerol (GLY), SORB, sucrose and NaCl at different concentrations. The treatment was followed by short-term incubation and determination of specific productivity of a reporter glucoamylase. We observed that the effect was largely dependent on the osmolyte used. The most beneficial was the addition of GLY and SORB, when the osmolality reached > 2.5 Osm kg−1. But since the correlation between a factor “osmolality” and a factor “specific activity” was low (r = 0.48), it was clear that more in-depth studies are necessary. Earlier, Yang et al. (2015) studied the total proteome of wild type Y. lipolytica subjected to 70 g L−1 of NaCl (Osm = 4.15 Osm kg−1) to reveal molecular background of erythritol (ERY) synthesis under these conditions. Using 2D-PAGE (two-dimensional polyacrylamide gel electrophoresis) followed by MS (mass spectrometry) of selected spots, the authors identified 44 differentially abundant proteins (DAPs) and determined their abundance level based on comparative band intensity. The same methodology was used in studies on dimorphic transition (Morín et al. 2007) and specificities of different amino acids (AAs) metabolism (Mansour et al. 2009) in Y. lipolytica.
Previously, we used global quantitative proteomics (liquid chromatography with tandem mass spectrometry; LC–MS/MS) to reveal molecular identities involved in synthesis of rose-like odor—2-phenylethanol (Celińska et al. 2015b). In the present research, we took advantage of this sensitive and unbiased method to study the impact of hyperosmolarity on synthesis of rs-Prots by Y. lipolytica. Considering our previous findings on the advantageous effect of decreased temperature on rs-Prots synthesis by this yeast species (Kubiak et al. 2021), we included this variant alone or in combination with hyperosmolarity to study its effect at a proteome level. Altogether, this research provides new insight into the actual relationship between hyperosmolarity and rs-Prots synthesis in Y. lipolytica characterized by biochemical, transcriptional and proteomic studies.
Materials and methods
Recombinant strain
Y. lipolytica strain GGY237 (Korpys-Woźniak et al. 2020) used in this study was a Po1h-derivative (MatA, ura3-302, xpr2-322, axp1-2) overexpressing a heterologous alpha-amylase-encoding gene from Sitophilus oryzae (GenBank: KP027641.1; (Celińska et al. 2015a)) under the synthetic 4UASpTEF promoter. The strain was constructed via Golden Gate strategy, as described previously (Celińska et al. 2017a, b).
Inoculum preparation
Precultures were developed from 15% GLY stocks stored at − 80 °C, plated on YPD agar medium containing [g L−1]: yeast extract (YE; Merck, USA), 10; bactopeptone (BP; Merck, Darmstadt, Germany), 20; glucose (POCH, Gliwice, Poland), 20; agar (BIOCORP, Warsaw, Poland), 20. Twenty-four-hour biomass was transferred into 1-L shake flasks containing 100 mL of YPG20 medium ([g L−1]: YE, 10; BP, 20; GLY, 20) and incubated at 30 °C with 250 rpm agitation in a rotary shaker (BIOSAN, ES-20, Riga, Latvia) for 23 h.
Bioreactor cultures and treatment conditions
Batch cultures were performed in Minifors 2 bioreactors (Infors, HT, Bottmingen-Basel, Switzerland) with a total volume of 2.6 L and a working volume of 1 L, in YPG120 medium, composed as follows [g L−1]: YE, 10; BP, 20; GLY, 120. The bioreactors were equipped with two Rushton turbines and sensors for temperature, pH and dissolved oxygen. pH was adjusted to 5.5 and controlled by automatic addition of 30% NaOH and 10% H2SO4. The temperature was maintained at 28 °C throughout the culture, except for the periods of thermal treatment, where indicated. Stirring and aeration were set at 700 rpm and 2 vvm, respectively. Antifoam agent (Antifoam 204, Sigma-Aldrich, St. Louis, MO, USA) was added automatically, when needed.
The treatments were executed starting from 20 h of culturing, which corresponds to the end of exponential/early stationary phase of growth. The cultures were conducted in four variants: (1) control (without treatment), (2; ↓Temp) with thermal treatment, (3; ↑Osm) with osmotic treatment, (4; ↓Temp↑Osm) with thermal and osmotic treatment (Fig. 1).
Treatment variants. Curves are based on actual measurements of temperature and osmolality read in real time by thermometer and in collected samples by osmometer, respectively. Y axis: temperature [°C]; auxiliary Y axis: osmolality [Osm kg−1], X axis: time [h]; 1) control variant, 2) ↓Temp variant, 3) ↑Osm variant, 4) ↓Temp↑Osm variant. Color code is explained in the legend. Given values are mean from biological quadruplicate ± SD
The applied thermal treatment (20 °C for 153.5 min) was previously optimized to maximize specific productivity of the target rs-Prot by Y. lipolytica (Kubiak et al. 2021). Hyperosmotic conditions (approx. 3.0–3.5 Osm kg−1) were also pre-optimized (not shown) and executed by aseptic addition of powdered SORB to reach the final concentration of 360 g L−1. Such manipulation does not transfer undesired microbiota to the culture, which was checked by surface plating (not shown). Noteworthy, at this concentration, SORB does not impact oxygen transfer rate, which was also verified. As demonstrated previously, SORB is not consumed by Y. lipolytica (Kubiak et al. 2019), and thus the implemented hyperosmolarity was stably maintained.
Each culture variant was conducted in four independent runs. Samples were collected throughout the culturing time, centrifuged (12,045 × g, 3 min; Eppendorf MiniSpin, Hamburg, Germany) and stored at − 20 °C (biomass and supernatant separately) to analyze the activity of the reporter protein, biomass accumulation, GLY consumption and synthesis of metabolites. Samples for total RNA and total protein isolation were collected at 23/25 h, centrifuged at 11,152 × g for 3 min at 4 °C (Eppendorf 5430 R, Eppendorf, Hamburg, Germany) and stored at − 80 °C until further analyzed. Each collected sample was analyzed in technical duplicate.
Enzymatic assay
Activity of the extracellular alpha-amylase (SoAMY) was analyzed according to a microSIT protocol described previously (Borkowska et al. 2019). One activity unit (AU) corresponds to the amount of an enzyme that triggers decrease in the starch-iodine staining value equivalent to 1 mg of starch per 1 mL, during 1 min at pH 5.0 and 40 °C, under applied experimental conditions.
Biomass concentration
Biomass concentration was determined using spectrophotometric measurements of absorbance at 600-nm wavelength (OD600). The biomass samples were first defrosted, washed twice in distilled water and suspended in water. All the samples were diluted 20–40 × , and 200 µL of them was transferred into a 96-well microplate. The absorbance was measured using a microplate reader (Multiskan Sky, Thermo Fisher Scientific Inc., Waltham, MA, USA). Subsequently, dry cellular weight (DCW) concentration was calculated according to a previously prepared standard equation (absorbance reading at 600 nm plotted against biomass concentration measured by a standard gravimetric method; gDCW L−1).
Osmolality
Osmolality of culture medium was determined by a freezing‐point depression method using an osmometer (Marcel os3000, Marcel, Zielonka, Poland). The thawed supernatants were appropriately diluted (due to high GLY or SORB concentration) and subjected to the measurement. One osmolality unit was defined as the quantity of osmoles of the osmotically active substance per kilogram of the sample [Osm kg−1].
Concentration of chemical compounds (high‐performance liquid chromatography, HPLC)
Concentration of small‐molecular weight metabolites including ERY, mannitol (MAN), citric acid (CA), alpha-ketoglutaric acid (aKG), externally added SORB and the carbon source (GLY) in the culturing medium was measured by HPLC. Defrosted and diluted supernatants were passed through 0.45-μm membrane syringe filters (Millipore, Merck-Millipore, Burlington, MA, USA). Analysis was conducted using a chromatograph VWR/Hitachi LaChrom Elite (Merck-Hitachi, Darmstadt, Germany), equipped with two detectors (RI L-2490 and UV L-2400) and a Rezex ROA 300 × 7.8 mm column (Phenomenex, Torrance, CA, USA), at 40 °C, under isocratic conditions, with a flow rate of 0.6 mL min−1 and 10 mM H2SO4, used as the mobile phase. Standard solutions of the chemicals were all purchased from Sigma-Aldrich (St. Louis, MO, USA). Quantitative analysis was performed in reference to the standard solutions (peak area) using EZChrom Elite (Agilent Technologies, Santa Clara, CA, USA) software.
Expression level of stress-related genes using RTqPCR
Total RNA isolation, reverse transcription (RT) reaction and real-time quantitative PCR (RTqPCR) were conducted as described by Borkowska et al. (2020). Briefly, Bead-Beat Total RNA Mini kit (A&A Biotechnology, Gdynia, Poland) and MixerMill MM400 (Retsch GmbH, Haan, Germany) were used for isolation of RNA. TranScriba Kit (A&A Biotechnology, Gdynia, Poland) and a Veriti Thermal Cycler (Applied Biosystems, USA) were used for the first cDNA strand synthesis. The RTqPCR was carried out using SYBR®Green PCR MasterMix kit B (A&A Biotechnology, Poland), FrameStar® 96 Well Semi-Skirted PCR Plates and a 7500 Real-time PCR Thermalcycler (Applied Biosystems, Foster City, USA). Real-time PCR primers targeting the intracellular stress-related genes and the internal calibrator were designed with Primer Expert Software (Applied Biosystems, Foster City, USA) (Supplemental Table S1). Comparative gene expression analysis of the treated samples was conducted vs the control culture (Fig. 1). Data analysis was carried out according to the delta-delta Ct (ΔΔCt) method (Livak and Schmittgen 2001).
Protein precipitation
The samples collected at 23/25 h of culturing were immediately centrifuged (as described above). The cellular pellets were resuspended in ice-cold breaking buffer (0.1 M sodium phosphate buffer, 5 μM DTT, 1 mM PMSF, 5% GLY) with glass beads (Sigma-Aldrich, St. Louis, MO, USA) and disrupted by repeated cycles (5 ×) of mixing at 30 strokes s−1 for 30 s in a MixerMill MM400 (Retsch GmbH, Haan, Germany) and incubation on ice for 1 min. The cellular debris were then separated by centrifugation (24,652 × g, 4 °C, 10 min; Eppendorf 5430 R; Eppendorf, Hamburg, Germany). The protein precipitation was carried out according to a standard methanol-chloroform method (Wessel and Flügge 1984). Briefly, the protein suspensions were combined with 4 volumes of methanol and 1 volume of chloroform, thoroughly mixed and centrifuged at 12,000 × g for 15 min. The aqueous phase was discarded, and organic phase was again mixed with 4 volumes of methanol, thoroughly mixed and centrifuged (12,000 × g, RT, 15 min). The supernatant was withdrawn, and the obtained protein pellets were air-dried. The protein pellet was resuspended in sterile 8 M urea/50 mM Tris–HCl/50 mM EDTA buffer (pH 8.0), according to Saito et al. (2019). The protein profile was examined for the protein integrity by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970). The protein samples were stored at − 80 °C until analyzed.
Tryptic digestion and LC–MS/MS analysis
The samples were processed through a filter-aided sample preparation (FASP) method (Wiśniewski et al. 2009) as described previously (Pinski et al. 2021) with minor modifications. The protein samples (50 µg) were brought to 200 µL with urea solution (8 M urea, 50 mM ammonium bicarbonate). Proteins were digested overnight with trypsin (Promega, Madison, WI, USA) at an enzyme to protein ratio of 1:50. About 1 µg of the resulting peptide mixture was injected on a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled with a nanoHPLC (UltiMate 3000 RSLCnano System, Thermo Fisher Scientific, Waltham, MA, USA). The parameters of the LC–MS/MS analysis were the same as in Pinski et al. (2021). Briefly, peptides were resolved on a 50 cm analytical column (AcclaimPepMapRLSC C18, Thermo Fisher Scientific; ID 75 μm, particle size 2 μm, pore size 100 Å) in the presence of 0.05% formic acid (JT Baker, Phillipsburg, NJ, USA) using a linear gradient of acetonitrile (2–40%) for 240 min. The mass spectrometric measurement was performed in the data-dependent mode using the Top12 method. The MS and MS/MS spectra were acquired at resolutions of 70,000 and 17,500, respectively. The performance of the LC–MS/MS platform was monitored using the QCloud quality control system (Chiva et al. 2018).
MS data processing and analysis
Raw proteomics data were processed using MaxQuant 1.6.7.0 with incorporated Andromeda search engine (Cox et al. 2011; Tyanova et al. 2016a) set to detect variable modifications (Oxidation—M, Acetyl—Protein N-term) and fixed modification (Carbamidomethyl—C). Identification was based on mapping to the UniProtKB Y. lipolytica reference proteome (accessed on 05.03.2021, 6454 entries). Default MaxQuant parameters were used. The false discovery rate (FDR) for the peptide and protein identification was set to 1%. Label free quantification (LFQ) was enabled. Statistical analysis was conducted using Perseus 1.6.15.0 (Tyanova et al. 2016b). Quality of data and overall profiles were analyzed by inspection of multi-scatter plots illustrating Pearson correlation, PCA analysis and hierarchical clustering of all the proteome profiles in quadruplicate. Differentially expressed proteins under the treatment conditions vs the control were defined by the results of Student’s t test, with correction based on permutation-based FDR (threshold FDR < 0.05 for each comparison). Protein filter was set at minimum 3/4 valid values in each group. Protein identification was restricted to at least 2 unique peptides. A protein was defined as DAP (differentially abundant protein) provided the fold change > 1.20 or < − 1.20. The mass spectrometry data were deposited to the ProteomeXchange Consortium (Vizcaíno et al. 2014) via the MassIVE repository with the dataset identifier PXD029106.
Bioinformatic tools and databases
Assignment of YALI identifiers, molecular functions and biological processes was done using GRYC (http://gryc.inra.fr/) and PANTHER databases (http://pantherdb.org/). Statistical overrepresentation test tool from PANTHER was used to indicate overrepresented classes of proteins within specified groups of up- and downregulated DAPs. Visualization of Venn diagrams was done using InteractiVenn (http://www.interactivenn.net/) online tool. The search for binding motifs within promoter regions of aa-tRNA synthetase-encoding genes was conducted using 500 bp regions upstream from the start codon and MEME online tool (https://meme-suite.org/meme/) with default settings (Bailey et al. 2015).
Statistical analysis
All the results are expressed as mean values ± standard deviation (± SD) of four biological replicates, each analyzed in at least duplicate. Statistical significance of the differences between compared sets of data was analyzed using one‐way analysis of variance (Microsoft Excel 2013 software). The level of significance was set at p < 0.05.
Results
Batch culture progression under the treatment conditions
Kinetics of batch cultivations in terms of GLY consumption, biomass formation and metabolites synthesis is given in Fig. 2. Until the treatment time, the cultures progression was highly corresponding in all the variants. Biomass productivity [dDCW dt−1] within this time range (0–20 h) was on average 1.35 g L−1 h−1. Following the treatment, the growth was largely slowed down. While in the control variant it continued up to 30 h reaching 1.25 g L−1 h−1, the exposure to ↓Temp decreased biomass productivity by more than half (equal to 0.45 g L−1 h−1 between 23 and 50 h). Increased osmolality of the culture medium led to further slowing down of growth (equal to 0.35 g L−1 h−1 between 23 and 50 h). The combined treatment ↑Osm↓Temp led to nearly complete arrest of biomass accumulation (equal to 0.07 g L−1 h−1 between 23 and 50 h). Until the treatment time, synthesis of metabolites was relatively low, and CA was the dominant molecule.
Growth (DCW), GLY utilization and metabolites synthesis over culturing time (Y axis and auxiliary Y axis: concentration in [g L−1]; X axis: time [h]) under different treatment variants and a control; a) control variant, b) ↓Temp variant, c) ↑Osm variant, d) ↓Temp↑Osm variant. Color code is explained in the legend. Abbreviations: GLY glycerol, ERY erythritol, MAN mannitol, CA citric acid, aKG alpha-ketoglutaric acid, DCW dry cellular weight, SORB sorbitol. Given values are mean from biological quadruplicate ± SD
Following the ↓Temp treatment, GLY consumption, ERY and MAN synthesis remained unchanged vs the control. On the other hand, CA synthesis and growth were substantially reduced when compared to the control (~ 45 vs ~ 71 g L−1 of CA at the end of culture, in the ↓Temp and control, respectively). The perturbation with ↑Osm (alone or in combination) induced multiple physiological reactions. Expectedly, synthesis of polyols (especially MAN) was significantly enhanced reaching > 40 g L−1 in the ↑Osm variant. ERY was synthesized up to > 20 g L−1 in both ↑Osm and ↑Osm↓Temp. For both polyols, some minor fluctuations in the concentration were observed, which is typical for Y. lipolytica cultures. Notably, GLY was not fully consumed until the end of any ↑Osm-treated culture.
In terms of the reporter rs-Prot synthesis and secretion, the implemented perturbations had varying effects (Fig. 3). Temperature downshift had immediate (within 3 h) positive impact on the rs-Prot synthesis and secretion, improving its titer [AU L−1] by 1.2-fold and specific activity [AU gDCW−1] by 1.6-fold (p < 0.001). Macroscopically visible response to the ↑Osm was observable after > 5 h. After that time, the cells started to restore growth, synthesis of metabolites and the rs-Prot. In terms of specific activity at this time point, no significant difference could be seen between ↑Osm and the control. On the other hand, some minor but statistically significant improvement was observed for the ↑Osm↓Temp variant (by 18%; p < 0.05; Fig. 3c). As could be observed in Fig. 3b, both ↑Osm-treated variants turned out to be beneficial for AU gDCW−1 parameter upon prolonged exposure. However, this observation results rather from limited growth, especially in the case of the ↑Osm condition (compare Figs. 2 and 3a). In the following time range (30 to 48 h), the ↑Osm↓Temp variant reached plateau in terms of extracellular activity (Fig. 3a) at the averaged level of 674.5 AU L−1 (vs 593.8 AU L−1 for the control, 703.2 and 593.4 AU L−1 in ↓Temp and ↑Osm, respectively).
Amounts of extracellular activity of the rs-Prot (SoAMY) under different treatment variants and the control, given as: a) activity (titer) in [AU L−1] in time; b) specific activity normalized per biomass in [AU gDCW−1] in time; c) specific activity in [AU gDCW−1] at a time point of gene expression analysis and proteomics samples collection (23/25 h). Y axis: extracellular activity given in [AU L−1] or [AU gDCW−1]; X axis: a, b) time [h]; c) treatment variant. Color code is explained in the legend. Results indicate mean values from quadruplicate repetitions ± SD. In (c) *, $ indicate statistical significance with variant C (*), ↓Temp ($) at p value < 0.05, $$ at p value < 0.01, ***, $$$ at p value < 0.001
Expression level of selected genes involved in temperature and hyperosmolarity stress response
Samples for molecular analyses were collected at the time point at which physiological response of the culture was observed (23 h for ↓Temp, and 25 h for ↑Osm and ↓Temp↑Osm). Gene expression analysis was conducted for a set of genes which, according to the literature, were shown responsive to either ↑Osm or ↓Temp in Y. lipolytica (Table 1). In addition, the heterologous gene encoding the reporter rs-Prot was also tracked for expression level. In contrast to what was seen macroscopically, the highest upregulation of the SoAMY gene was detected under ↑Osm (Fig. 4). Likewise, a number of the stress-related genes, including those involved in gene expression regulation (Skn7 and Sko1), folding and maintenance in folding-competent state (DnaJ, Fmo1, HSPs), but also in synthesis of compatible solutes (Tpss, Gcy12) were significantly upregulated upon ↑Osm. ↓Temp treatment led to significant enhancement in SoAMY expression but negligible impact on expression of the stress response genes. Expression pattern of the genes was somehow similar under ↑Osm↓Temp as under ↑Osm, but in majority of cases the upregulation level was diminished due to the decreased temperature. Collectively, based on these data, it could be concluded that ↑Osm treatment imposed actual stress to the producer cells and awoke a transcriptional response. On the other hand, ↓Temp attenuated this response in the ↑Osm↓Temp variant. Sole ↓Temp did not impose any transcriptional reaction of the analyzed gene targets but enhanced expression of the heterologous gene under the synthetic promoter.
Expression level of selected genes analyzed by RTqPCR. Samples for gene expression analysis were collected at 23/25 h of culturing. Y axis: relative quantitation level determined by ΔΔCt analysis; X axis: analyzed genes—shortened name and YALI code. RQ relative quantitation. Shared/specific molecular functions of the genes are indicated. Color code is explained in the legend. Error bars indicate relative quantitation (RQ) ± SD from technical duplicates
Treatment-induced proteomic changes—phenomena shared between the treatment variants
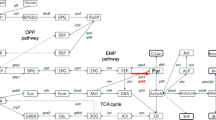
Gene expression profiles did not explain macroscopic observations. Therefore, in the next step, we conducted high-throughput quantitative proteomics analysis to get more insight into molecular bases of the observed physiological responses. The analysis was conducted on biological quadruplicate samples. The replicates showed high level of Pearson correlation, and they were grouped together in PCA analysis and in hierarchical clustering (Supplemental Table S2). On average, 2330 ± 93 proteins were identified in each sample (median: 2339). DAPs were indicated based on Student’s t test and FDR correction for multiple comparisons. DAPs were then filtered for FC > 1.20 or < − 1.20. The applied data processing pipeline yielded no DAPs upon comparison of ↓Temp vs control. Implementation of ↑Osm alone yielded 63 downregulated and 49 upregulated DAPs, while in combination (↑Osm↓Temp) resulted in 42 downregulated and 11 upregulated DAPs. Detailed lists of all the DAPs with functional annotation and fold change (FC) of abundance are given in Supplemental Table S3. Identification of common and unique DAPs responsive to ↑Osm and ↑Osm↓Temp was conducted by Venn diagram analysis. The results are shown in Fig. 5, and a detailed list of the members of each specific group is given in Supplemental Table S4. Among the uniformly upregulated DAPs, we identified three aldo–keto reductases (AKRs), including erythrose reductases Gcy12 (B07117) and Gcy13 (A15906). The commonly downregulated DAPs covered i.a. proteins involved in glycolysis (Pyc1/C24101), tricarboxylic acid cycle (TCA) (Acl1/E34793, Acl2/D24431), fatty acid (FA) metabolism (Fas1/B15059, Fas2/B19382, Pox3/D24750, Aal7/E20405), and proteins involved in proteolysis (F20592, C10494), AAs metabolism (B21846, A21417, D25168, C22088) and translation (a group of aa-tRNA synthetases, isoleucyl-/A00264, prolyl-/E05027, asparaginyl-/E05005).
The set of upregulated DAPs shared between ↑Osm and ↑Osm↓Temp (5 DAPs) was significantly enriched in molecular function alditol:NADP + 1-oxidoreductase activity, represented by 3 DAPs (per 13 total in the Y. lipolytica reference proteome; enrichment fold (EF) > 100). No significant enrichment in biological process was identified in this set of DAPs. The set of downregulated DAPs shared between ↑Osm and ↑Osm↓Temp (19 DAPs) was significantly enriched in several biological processes, including: FA biosynthetic (4/20; EF > 100) and metabolic processes (5/58; EF: 29.26), transsulfuration (2/6; EF: > 100), acetyl-CoA biosynthetic process (2/7; EF: 96.98) and tRNA aminoacylation for protein translation (3/35; EF: 29.09). Shared, downregulated molecular functions were enriched in ATP citrate synthase activity (2/2; EF: > 100), fatty-acyl-CoA synthase activity (2/2; EF > 100) and FA synthase activity (2/6: EF: > 100), aminoacyl-tRNA ligase activity (3/35; EF: 29.09) as well as small molecule (10/804; EF: 4.22) and anion binding (9/746; EF: 4.09). Specificities of unique proteomes awaken by ↑Osm and ↑Osm↓Temp are described in the following sections (Quantitative proteome of ↑Osm-treated Y. lipolytica vs control and Quantitative proteome of ↓Temp↑Osm-treated Y. lipolytica vs control). Selected DAPs with their abundance profiles averaged across all the samples, under all the treatment variants, are presented in a form of heat map (Fig. 6).
Heat map of selected DAPs. Up or down-representation of a given protein is color-coded according to the provided legend based on log2(LFQ Intensity) mean value from quadruplicate. Columns represent specific treatment conditions indicated at the top of the heat map. Rows represent individual DAPs indicated on the right site of the heat map. The heat map was prepared using Morpheus online tool (https://software.broadinstitute.org/morpheus/). Specific DAPs are given with their shortened name, function and shortened YALI code
Quantitative proteome of ↑Osm-treated Y. lipolytica vs control
Implementation of high SORB-induced hyperosmolarity stress alone yielded 112 DAPs. The proteins were up-/down-regulated by a maximum fold factor of 11.08-/-3.16-FC (Supplemental Table S3). Functional annotation of DAPs was cross-referenced in PANTHER, GRYC and NCBI (via blastp similarity search), resulting in a carefully inspected landscape of biological processes and molecular functions deregulated under ↑Osm. PANTHER-based functional annotation and statistical overrepresentation analyses (Fig. 7) indicated that protein refolding was the dominant upregulated biological process. The refolding process was executed by the action of cytosolic (Ssa6/E35046, Ssa8/D22352, Hsp104/E27962, Hsp90/B15840), ER-localized (E25696, F00880) and mitochondrial (Hsp78/F12463; ISU1/B04928) (co-)chaperones induced by the factor of 1.23 to 3.8 (Fig. 6, Supplemental Table S3). In addition, oxidoreductases, incl. AKRs involved in polyols synthesis, were among the most upregulated DAPs (Gcy13/A15906, Gcy12/B07117, A19910, F24937, D08778), reaching average upregulation of > fourfold (compare Fig. 6). The other upregulated DAPs of high-relevance to the awaken stress response are proteins involved in oxidative stress response (glutathione peroxidase/E02310, glyoxalase/F00682), glycogen phosphorylase involved in mobilization of stored glycogen (F04169), highly upregulated proteins (> 2-FC) involved in mitochondrial respiration (D08602, F24409, D09933). Interestingly, DAPs involved in cellular membrane invagination and endocytosis (Pil1/C11341, D13442), vesicle transportation (B14102, F27379) and the major vacuolar protease Prb1 (A06435) were all upregulated, suggesting specific cellular reaction.
Significantly enriched biological processes (Biol. Proc.) and molecular functions (Mol. Func.) within a set of DAPs deregulated in response to ↑Osm. Only main, selected processes and functions are shown. Analysis was conducted with PANTHER statistical overrepresentation tool by feeding a list of DAPs’ YALI names. Numbers in brackets indicate the number of DAPs assigned to that group; the following numbers indicate enrichment fold
Molecular functions and biological processes downregulated under ↑Osm were primary represented by decreased abundance of amino acid-tRNA (aa-tRNA) synthetases specific to isoleucine (A00264), lysine (F16291), proline (E05027), leucine (E24607), asparagine (E05005), glutamate (E28468) and tryptophan (B08943) (Figs. 6 and 7). Likewise, proteins involved in ribosome biogenesis (E31625, F12661) and biosynthesis of AAs (Aro10/D06930, Bat2/F19910, Pro3/B14399), including sulfur compounds (CysK-Met25/D25168, MetB/C22088, B14509) were also identified within this group, altogether indicating significant attenuation of protein synthesis. In this regard, translation elongation factor gamma (eEF-1) was among the most downregulated DAPs under ↑Osm (D16467 FC − 1.63; and its homolog B12562 FC − 1.19). Furthermore, three karyopherins, involved in nuclear im-/export, were identified within this group of DAPs (E16324, F10098, D12144; Supplemental Table S3). The set of ↑Osm-driven downregulated DAPs was enriched in proteins representing several metabolic pathways involved in central carbon metabolism: FA metabolism (Fas1/B15059, Fas2/B19382, Fao1/B14014, Aal7/E20405, Pox3/D24750), glycolysis and TCA (Pyc1/C24101, Pyk1/F09185, Pgi/F07711, Acl1/E34793, Acl2/D24431). While not significantly enriched in the overrepresentation analysis (Fig. 7), several genes involved in cytoskeleton formation and remodeling were significantly downregulated in response to ↑Osm, including actin capping protein (C06325), tubulins (Tub1/E19360, Tub2/E00726) or a protein promoting actin folding (D13090). Within the downregulated DAPs, we also identified several proteins involved in proteolysis via vacuolar (C10494, F20592) or proteasomal (F28413, B02860, F09834) degradation.
Quantitative proteome of ↓Temp↑Osm-treated Y. lipolytica vs control
Application of hyperosmolarity in combination with decreased temperature yielded 53 DAPs, which is less than half of what was identified for ↑Osm-treated culture. This observation suggests that additional execution of the low temperature treatment diminished impact of ↑Osm. The proteins were up-/down-regulated by a maximum fold factor of 13.3-/-1.91-FC (Supplemental Table S3). PANTHER-based functional annotation and statistical overrepresentation analyses (Fig. 8) rendered no significantly enriched biological processes within these upregulated DAPs. On the other hand, significantly enriched molecular function was represented by AKR activities, including Gcy12 (B07117) and Gcy13 (A15906), but also A19910 and D12386 (see also Fig. 6). Interestingly, within DAPs upregulated in response to ↓Temp↑Osm, we identified Atg8 (E02662), which is a multifunctional protein involved in autophagy-related processes, including selective nucleophagy, mitophagy, endoplasmic reticulum-specific autophagic process, autophagosomes formation and expansion, membrane invagination and filamentous growth regulation.
Significantly enriched biological processes (Biol. Proc.) and molecular functions (Mol. Func.) within a set of DAPs deregulated in response to ↑Osm↓Temp. Only main, selected processes and functions are shown. Analysis was conducted with PANTHER statistical overrepresentation tool by feeding a list of DAPs YALI names. Numbers in brackets indicate the number of DAPs assigned to that group; the following numbers indicate enrichment fold
Molecular functions and biological processes downregulated under ↑Osm↓Temp were enriched in DAPs related to attenuation of protein synthesis. Primarily, we observed significantly decreased representation of aa-tRNA synthetases, specific to isoleucine (A00264), proline (E05027), arginine (E17985), asparagine (E05005) (downregulated by > 1.2-fold; Fig. 6, Supplemental Table S3). Furthermore, there was an additional panel of significantly downregulated aa-tRNA synthetases that did not meet the criterion of 1.2-FC (specific to aspartate (D22264), phenylalanine (E22979), methionine (F29843); not shown). Next, biosynthesis of several AAs was significantly enriched within these downregulated DAPs, represented by 4HPPDase/B21846 involved in tyrosine metabolism, Aat2/F29337 and Glt1/B19998 implicated in glutamate synthesis, glutamine synthetases /D13024 and A20108, and Arg4/D26367, CysK-Met25/D25168, MetB/C22088, playing roles in synthesis of arginine and sulfur-containing AAs (Fig. 6, Supplemental Table S3). As in the case of ↑Osm-induced response, the ↑Osm↓Temp proteome was characterized by significant downregulation of FA metabolism (Fas1/B15059, Fas2/B19382, Fao1/B14014, Aal7/E20405, Pox3/D24750), glycolysis and TCA (Pyc1/C24101, 6-phosphofructo-2-kinase/D21010, Acl1/E34793, Acl2/D24431). The majority of these DAPs were shared between the ↑Osm-downregulated set of proteins (Fig. 6, Supplemental Table S3).
aa-tRNA synthetases promoter regions sequence search for common binding motif
Clearly marked downregulation of several aa-tRNA synthetases under ↑Osm and ↑Osm↓Temp treatments led us to hypothesis that maybe genes encoding these ligases are co-regulated. To test this, we extracted ~ 500 bp of genomic DNA regions upstream the ATG codon and analyzed them for occurrence of specific binding motifs. The results of this analysis are shown in Fig. 9. No single motif, uniformly present in all eight analyzed regions could be identified. The most frequent motif was #4 TGACTCAY (7/8), followed by #1 GRCTTTTTTTKTHTKKWTTTT (6/8) and #3 GGGWKGAAAAA (6/8).
Visual representation of MEME motif search results conducted on 500 bp DNA sequence of genomic DNA upstream start codon of specific aa-tRNA synthetases identified in this work as downregulated DAPs. Name indicates specificity of a given ligase: isoleu/A00264, lys/F16291, pro/E05027, leu/E24607, asp/E05005, glu/E28468, tryp/B08943, arg/ E17985. Identified motifs are color coded according to the provided legend
Discussion
Synthesis, maturation and secretion of rs-Prot is a complex biological process relying on multiple molecular functions. Depending on the biochemical characteristics of the rs-Prot and physiology of the host, different steps of the process may be limiting (Delic et al. 2013; Korpys-Woźniak et al. 2020; Swietalski et al. 2020). Nevertheless, high provision of HSPs (chaperones and foldases) and osmolytes within the producer cell seems to be a potentially generalizable strategy for enhancing rs-Prot production in bacteria and yeast (Oganesyan et al. 2007; Lazar et al. 2011). It is a widely recognized mechanism, that under exposure to different environmental stress factors, the cell increases the intracellular abundance of the protective agents to preserve delicate proteins and membranes from denaturation. This mechanism, among others, found survival and tolerance under adverse conditions. Development of a tolerant phenotype relies on active remodeling of cellular organization and thus requires significant transcriptional and translational effort. For example, osmotolerance in Saccharomyces cerevisiae is not reliant on sole intracellular accumulation of GLY, since osmotically conditioned cells washed free of GLY retain their capacity for growth on high salinity media. In contrast, blockage of protein synthesis inhibits the acquisition of the tolerant state, indicating a protein-mediated phenotype (Blomberg and Adler 1989; Blomberg 1995). Upregulation of HSP abundance is a part of general stress response, awaken under different stress conditions. Since HSPs execute their folding/protective role in response to stress conditions and as a part of a “regular” biosynthesis process, they constitute a link between the stress response and rs-Prots synthesis. The primary aim of this research was two-fold: (1) to settle if prolonged exposure to hyperosmolarity enhances synthesis of the rs-Prots in Y. lipolytica, (2) to get an insight into molecular background of the macroscopically observed physiological reaction of the cell to ↑Osm/↓Temp/↑Osm↓Temp treatments. Previously, we optimized thermal treatment conditions for enhanced synthesis of rs-Prots by Y. lipolytica (Kubiak et al. 2021). In the present study, we implemented that optimized treatment alone (↓Temp) or in combination with hyperosmolarity (↑Osm↓Temp). We were interested if application of the two treatment factors, presumably beneficial for rs-Prot synthesis, may exert combined/contradictory action, and what is the molecular background of it.
We induced the hyperosmotic conditions in Y. lipolytica batch cultures by abrupt addition of a non-utilizable by this yeast chemical compound—SORB. Such an approach allowed to stably maintain the stress conditions. In addition, since SORB is also inert for the rs-Prot, the risk of inactivating the reporter protein was eliminated. The ↑Osm stress was executed at the end of exponential growth phase. The treatment was followed by temporary growth arrest (3–5 h; Fig. 2), while the samples for molecular analyses were collected when the growth recommenced. Earlier studies showed that this growth-less adaptation stage is highly dynamic in terms of proteome remodeling, but the absolute rate of total protein synthesis is then roughly fourfold decreased (Blomberg 1995). In the present study, we were more interested in practical consequences of the newly developed organization of the cell that could potentially contribute to enhanced rs-Prot synthesis. Hence, it was reasonable to address this issue by analyzing the cells actively producing proteins after the recommencement of growth. For details on dynamic changes in the yeast osmo-proteome directly after osmotic shock (within 1 h), the reader is referred to excellent works by Blomberg (1995) and Soufi et al. (2009).
Considering the literature data, the observed growth arrest (3–5 h directly after the treatment execution) was expected in terms of occurrence, intensity and duration. The primary mediator of the osmostress response in yeast—Hog1—is known to regulate delay in the cell cycle to permit stress adaptation prior to its further progression (Clotet and Posas 2007). S. cerevisiae exposed to 1.4 Osm kg−1 by NaCl recovered growth within 1 h (Blomberg 1995). Here, we implemented over twofold higher osmolarity which had a significant limiting effect on the growth rates. In terms of metabolites synthesis, ↓Temp led to ~ 60% decrease in final titers of CA vs control, but the other typical compounds remained at unchanged levels. The same relates to GLY utilization profile shared between the ↓Temp and the control conditions. In this case, even though growth rate was significantly reduced, GLY was consumed at similar rates. Therefore, considering the observed physiological reaction of the cells, i.e. similar GLY consumption profile accompanied by limited growth and CA formation, but significantly elevated amounts of rs-Prot (Fig. 3), we infer that ↓Temp allows to channel carbon flux toward the rs-Prot synthesis. On the other hand, our proteomics data did not render any specific protein that was differentially abundant between the two conditions and could be potentially implicated in this redirection. This suggests that it was not regulated at proteomics level. Likewise, the gene expression analysis (Fig. 4) did not indicate any specific gene that could contribute to the enhanced quantities of the rs-Prot produced under ↓Temp. This indicates that these reactions must be regulated at some other than transcriptional level, or that some other genes are implicated, beyond the set of genes analyzed here.
Cultures subjected to ↑Osm (or ↑Osm↓Temp) were characterized by significantly elevated amounts of MAN (7x/ 8 × vs control) and ERY (2 × vs control), as expected. Proteomics and transcriptional data show that erythrose reductases Gcy12 and Gcy13, belonging to AKRs, were the key molecular identities, sharply responding to ↑Osm with enhanced expression. The omics data revealed several other AKR-type reductases (A19910, F24937, D08778) upregulated in response to ↑Osm, whose potential implication in the polyols synthesis could be verified in the following studies. Considering the very high disproportion in metabolic profiles developed under the control and the ↑Osm-treated variants, we looked for molecular bases of the metabolic fluxes redistribution. Apart from significant upregulation of the AKRs, we observed concerted downregulation of proteins involved in glycolysis, TCA and FA synthesis. Under hyperosmolarity, S. cerevisiae increases carbon fluxes toward GLY at the expense of carbon dioxide production (Blomberg 1995; Soufi et al. 2009). It was proposed that downregulation of glycolysis and ethanol/carbon dioxide synthesis is one of the prerequisites for increased formation of GLY, required for survival under the stress conditions in S. cerevisiae (Blomberg 1995; Soufi et al. 2009). The second prerequisite is transcriptional upregulation of GPD1 (glycerol-3-phosphate dehydrogenase), the key enzyme required for GLY synthesis. It seems that a very similar mechanism operates in Y. lipolytica: (i) upregulation of the key enzymes involved in the polyols synthesis, which are known to be regulated at transcriptional level by ↑Osm (Trassaert et al. 2017), accompanied by (ii) downregulation in the other carbon fluxes, including glycolysis, TCA and FA biosynthesis. Proteins directly involved in this mechanism in Y. lipolytica were revealed and are indicated in Fig. 6 and Supplemental Table S3. Corresponding data regarding downregulation of glycolysis and TCA proteins (including Eno2, isocitrate dehydrogenase, Sdh1) and upregulation of AKRs were reported previously for salt-induced ↑Osm in Y. lipolytica, based on 2D-PAGE global proteomics (Yang et al. 2015).
In contrast to what was previously postulated (Fiedurek 1998; Oganesyan et al. 2007; Lazar et al. 2011; Kubiak et al. 2019), we did not observe any direct causative relationship between ↑Osm and ↑rs-Prot titer in Y. lipolytica (Fig. 3), at least when SORB was used as the osmolarity inducer at 3 Osm kg−1. The key modulator of osmo-stress response (Hog1) was not deregulated under ↑Osm/↑Osm↓Temp when compared to the control, at neither transcriptional (Fig. 4) nor protein level (Fig. 6, Supplemental Table S3). The same was reported previously with ↑Osm-treated Y. lipolytica studied by 2D-PAGE (Yang et al. 2015). It was expected, since the components of MAPK pathway are mainly responding to a specific inducer by their phosphorylation status rather than at transcriptional level (Soufi et al. 2009). Hence, considering a typical physiological reaction and some molecular evidence pointed hereafter, we speculate that Hog1 was indeed activated under the ↑Osm treatment, and its signaling cascade was initiated. An example of such a molecular evidence is high upregulation of SKO1 (Fig. 4) encoding a transcription factor that is a direct downstream target of Hog1 (Gomar-Alba et al. 2013). Sko1 is an ATF/CREB repressor which is a substrate of Hog1 and is phosphorylated under osmotic stress. Such mechanism exemplifies one of nuclear/transcriptional functions of Hog1, executed via the Sko1 repressor. Transcriptional functions of Hog1 are also executed by several transcriptional activators, including Msn2/4, Smp1 and Hot1. Except for that, Hog1 also plays a cytoplasmic/non-transcriptional role in promoting stress tolerance by sequential activation of multiple targets. One of such targets is the Rck2 serine-threonine protein kinase (Teige et al. 2001). Since it does not respond to ↑Osm at transcriptional level, we have not seen it as DAP; however, our data suggest that we observed downstream physiological reaction of the Rck2’s activation. As proved by Teige et al. (2001), Hog1-driven activation of Rck2 is responsible for attenuation of protein synthesis in response to osmotic challenge through modification of translation elongation factor 2 (EF-2). It was shown that Rck2 regulates translation by phosphorylation of EF-2, leading to its decreased binding affinity to ribosomes and consequently to a lower level of protein synthesis (Teige et al. 2001). In the present study, the ↑Osm treatment decreased both titer and specific activity of the target rs-Prot (Fig. 3), although its gene expression was significantly increased (Fig. 4). These contradictory data from transcriptional and proteomics analysis are even further strengthened by significant upregulation of several HSPs and chaperones, which are known to assist protein synthesis, folding and maturation in Y. lipolytica (Celińska and Nicaud 2019). In addition, our recent study clearly indicated that co-overexpression of Ssas genes, identified as DAPs under ↑Osm (Figs. 4, 6, 7, Supplemental Table S3), leads to significant increase in titers of overproduced rs-Prots (Korpys-Woźniak et al. 2021). Here, even upon upregulation of the Ssas and the other chaperoning activities, the final yield of the rs-Prot under ↑Osm was lower. For most of eukaryotic organisms, the rapid inhibition of protein synthesis is an element of a stress response (Blomberg 1995; Simpson and Ashe 2012). As shown by Dunand-Sauthier et al. (2005) global protein synthesis is rapidly, but transiently, reduced in S. cerevisiae following exposure to osmotic stress, induced by KCl. Within 1 h, the cells restore normal global protein synthesis. Teige et al. (2001) showed that this inhibition is driven by Hog1-Rck2 cascade activation. Our data revealed several specific downstream mechanisms by which this downregulation of protein synthesis is executed. Primarily, we observed massive downregulation of aa-tRNA synthetases (Fig. 6, Supplemental Table S3). Their downregulation limits translation and renders the already transcribed tRNAs uncharged. The uncharged tRNAs are known potent regulators of both transcription (by direct interaction with promoter regions) and translation (by direct interaction with translation machinery) (Raina and Ibba 2014; Gomez and Ibba 2020). Except for downregulation of the aa-tRNA synthetases in both ↑Osm and ↑Osm↓Temp, we observed systemic downregulation of proteins involved in AAs biosynthesis, and downregulation of translation elongation factor gamma (its two homologs—D16467, B12562) (Fig. 6, Supplemental Table S3). This elongation factor could work as an interaction partner with uncharged tRNA. In addition, Hog1 is known to physically interact with Sfp1, the transcription factor that regulates ribosome biogenesis. Phosphorylated Sfp1 is recruited to the ribosome biogenesis genes (Sellam et al. 2019), but this mechanism is abolished under stress. Among DAPs, we identified several proteins significantly downregulated in response to ↑Osm and ↑Osm↓Temp which could be downstream consequences of Hog1-Sfp1 interaction. Based on all these observations, we infer that ↑Osm conditions decrease proteins biosynthesis in Y. lipolytica cells by several concerted mechanisms, including: (i) downregulation of aa-tRNA synthetases and (ii) translation elongation factor gamma, (iii) downregulation of AAs biosynthesis and (iv) ribosomes biogenesis. We speculate that the positive impact of the increased abundance of chaperones, HSPs and osmolytes does not outbalance the losses by the former phenomena. The observed proteomics profiles suggest that de novo synthesis of proteins is arrested, but the already synthesized polypeptides are subjected to chaperoning/folding activity. Similar patterns of elevated abundance of HSPs, including cytoplasmic Ssas, severe downregulation of translation elongation factor alpha and partial downregulation of genes involved in AAs and ribosomes biosynthesis, were observed in the formerly reported proteomics profile of osmostressed Y. lipolytica (Yang et al. 2015).
We found the concerted downregulation of so many aa-tRNA synthetases very intriguing. As a side question posed here, we asked if these genes can be co-regulated by sharing a common binding motifs in their regulatory regions. Furthermore, we were interested if these potential motifs are known to be specific to any of known transcription factors inflicted in stress response. Our analysis conducted on promoter regions (Fig. 9) revealed several motifs potentially driving the putative co-regulation. One of the most interesting was a motif TGACTCAY, present in 7 out of 8 analyzed regions. The motif shares high similarity with the ATF/CREB consensus sequence TGACGTCA (Rep et al. 2001), specific to i.a. Sko1, that was upregulated at transcriptional level in the present study (Fig. 4). While it is still highly speculative, the downregulated aa-tRNA synthetases could be the trans-targets of the TGACTCAY-specific transcription factors acting within osmostress response. Some DNA motifs potentially directing this regulation are shown in Fig. 9, to be further studied through detailed biochemical experimentation.
Another aspect of the awaken osmostress response observed in this study was localized to plasma membrane and cell wall. Among the upregulated DAPs responsive to ↑Osm, we found several proteins involved in eisosomes formation, membrane invagination and endocytosis, including Pil1 (C11341). Pil1 is the key component of eisosomes—specific structures distributed uniformly across the cell surface periphery, serving as endocytic hotspots. Considering the growth arrest that we observed in the cultures following ↑Osm/↑Osm↓Temp execution, the increased endocytosis was rather surprising. Therefore, we speculate that the increased endocytosis could be related to sequestration of membrane channels and transporters, which were targeted for degradation in the vacuole. What supports our statement is that: (i) Hog1 migrates to the cell periphery under ↑Osm, to close Fps1 aquaglyceroporin (Lee et al. 2013), (ii) AAs transporters are redundant at the cellular membrane under ↑Osm, as the yeast cells respond with depression of AA uptake after osmotic shock (Blomberg 1995), (iii) eisosomes are regulatable portals that govern both location and magnitude of membrane traffic into the cell; though it is not known if the elements building eisosomes are directly interacting with Hog1, (iv) among the upregulated DAPs we identified several vacuolar proteases, including the major vacuolar protease Prb1 (A06435). Results from 2D-PAGE followed by MS (Yang et al. 2015) showed that indeed upon ↑Osm one of the key potassium ion channels was severely downregulated, demonstrating its redundancy under the treatment. Likewise, upregulation of Prb1 (called there cerevisin) was observed in that study, which corresponds to our current observations. It would be very interesting to test this hypothesis by more detailed molecular biology research.
Finally, the proteomic profiles generated under ↑Osm suggest significant remodeling in the cytoskeleton and cell wall. Significant upregulation of UTR2 cell wall protein (B15510) indicates remodeling taking place at the cell wall, enhancing its rigidity. Soufi et al. (2009) showed that the cell wall integrity pathway is activated under osmostress by phosphorylation of Pkh1 and Ypk1/Ypk2 involved in maintenance of cell wall integrity during environmental stresses. In addition, we observed a number of DAPs down-regulated in response to ↑Osm that are involved in actin cables folding and actin-based transportation. As discussed earlier, increased external osmolarity causes a rapid loss of actin filament cables (Blomberg 1995). It was even suggested by that author that actin filament organization may act as an osmosensor responding to changes in turgor pressure and rapidly mediating this response to the general control points of the cell cycle. In relation to that, upregulation of Sko1, detected at transcriptional level in this study (Fig. 4), is known to limit filamentation (Su et al. 2013). Our microscopic observations of the cells following the ↑Osm treatment showed that the cells represented mainly ovoid/elongated ovoid morphotype (not shown).
In conclusion, we studied physiological, transcriptional and proteomic response of recombinant Y. lipolytica strains overproducing a heterologous rs-Prot subjected to ↑Osm, ↓Temp and ↑Osm↓Temp treatments. Our results indicate that hyperosmolarity is detrimental for the synthesis of rs-Prots, and molecular bases of this observation are described. Key molecular players involved redirection of carbon fluxes from CA and growth to polyols were revealed through the global proteomics analysis.
Availability of data and material
All data accompanying this research are presented directly in the manuscript and supplementary materials. The mass spectrometry data were deposited to the ProteomeXchange Consortium via the MassIVE repository with the dataset identifier PXD029106.
Code availability
Not applicable.
References
Aguilera J, Randez-Gil F, Prieto JA (2007) Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol Rev 31:327–341. https://doi.org/10.1111/j.1574-6976.2007.00066.x
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME Suite. Nucleic Acids Res 43:W39–W49. https://doi.org/10.1093/nar/gkv416
Blomberg A (1995) Global changes in protein synthesis during adaptation of the yeast Saccharomyces cerevisiae to 0.7 M NaCl. J Bacteriol 177:3563–3572. https://doi.org/10.1128/jb.177.12.3563-3572.1995
Blomberg A, Adler L (1989) Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J Bacteriol 171:1087–1092. https://doi.org/10.1128/jb.171.2.1087-1092.1989
Borkowska M, Białas W, Kubiak M, Celińska E (2019) Rapid micro-assays for amylolytic activities determination: customization and validation of the tests. Appl Microbiol Biotechnol 103(5):2367–2379. https://doi.org/10.1007/s00253-018-09610-0
Borkowska M, Białas W, Celińska E (2020) A new set of reference genes for comparative gene expression analyses in Yarrowia lipolytica. FEMS Yeast Res 20(7):foaa059. https://doi.org/10.1093/femsyr/foaa059
Celińska E, Białas W, Borkowska M, Grajek W (2015a) Cloning, expression, and purification of insect (Sitophilus oryzae) alpha-amylase, able to digest granular starch, in Yarrowia lipolytica host. Appl Microbiol Biotechnol 99:2727–2739. https://doi.org/10.1007/s00253-014-6314-2
Celińska E, Borkowska M, Białas W (2017a) Enhanced production of insect raw-starch-digesting alpha-amylase accompanied by high erythritol synthesis in recombinant Yarrowia lipolytica fed-batch cultures at high-cell-densities. Process Biochem 52:78–85. https://doi.org/10.1016/j.procbio.2016.10.022
Celińska E, Borkowska M, Białas W, Korpys P, Nicaud JM (2018) Robust signal peptides for protein secretion in Yarrowia lipolytica: identification and characterization of novel secretory tags. Appl Microbiol Biotechnol 102:5221–5233. https://doi.org/10.1007/s00253-018-8966-9
Celińska E, Ledesma-Amaro R, Larroude M, Rossignol T, Pauthenier C, Nicaud JM (2017b) Golden Gate Assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica. Microb Biotechnol 10:450–455. https://doi.org/10.1111/1751-7915.12605
Celińska E, Nicaud J-M (2019) Filamentous fungi-like secretory pathway strayed in a yeast system: peculiarities of Yarrowia lipolytica secretory pathway underlying its extraordinary performance. Appl Microbiol Biotechnol 103:39–52. https://doi.org/10.1007/s00253-018-9450-2
Celińska E, Olkowicz M, Grajek W (2015b) L-Phenylalanine catabolism and 2-phenylethano synthesis in Yarrowia lipolytica-mapping molecular identities through whole-proteome quantitative mass spectrometry analysis. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fov041
Chang CC, Ryu DDY, Park CS, Kim JY (1998a) Improvement of heterologous protein productivity using recombinant Yarrowia lipolytica and cyclic fed-batch process strategy. Biotechnol Bioeng 59:379–385. https://doi.org/10.1002/(SICI)1097-0290(19980805)59:3%3c379::AID-BIT15%3e3.0.CO;2-9
Chang CC, Ryu DDY, Park CS, Kim JY, Ogrydziak DM (1998b) Recombinant bioprocess optimization for heterologous protein production using two-stage, cyclic fed-batch culture. Appl Microbiol Biotechnol 49:531–537. https://doi.org/10.1007/s002530051209
Chiva C, Olivella R, Borràs E, Espadas G, Pastor O, Solé A, Sabidó E (2018) QCloud: a cloud-based quality control system for mass spectrometry-based proteomics laboratories. PLoS ONE 13:e0189209. https://doi.org/10.1371/journal.pone.0189209
Clotet J, Posas F (2007) Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol 428:63–76. https://doi.org/10.1016/S0076-6879(07)28004-8
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. https://doi.org/10.1021/pr101065j
Delic M, Valli M, Graf AB, Pfeffer M, Mattanovich D, Gasser B (2013) The secretory pathway: exploring yeast diversity. FEMS Microbiol Rev 37:872–914. https://doi.org/10.1111/1574-6976.12020
Dunand-Sauthier I, Walker CA, Narasimhan J, Pearce AK, Wek RC, Humphrey TC (2005) Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot Cell 4:1785–1793. https://doi.org/10.1128/EC.4.11.1785-1793.2005
Fiedurek J (1998) Enhancement of β-galactosidase production and secretion by high osmotic stress in Penicillium notatum. Microbiol Res 153:65–69. https://doi.org/10.1016/S0944-5013(98)80023-2
Flores CL, Gancedo C, Petit T (2011) Disruption of Yarrowia lipolytica TPS1 gene encoding trehalose-6-P synthase does not affect growth in glucose but impairs growth at high temperature. PLoS ONE. https://doi.org/10.1371/journal.pone.0023695
Gasmi N, Ayed A, Ammar BBH, Zrigui R, Nicaud JM, Kallel H (2011a) Development of a cultivation process for the enhancement of human interferon alpha 2b production in the oleaginous yeast, Yarrowia lipolytica. Microb Cell Fact. doi: https://doi.org/10.1186/1475-2859-10-90
Gasmi N, Ayed A, Nicaud JM, Kallel H (2011b) Design of an efficient medium for heterologous protein production in Yarrowia lipolytica: case of human interferon alpha 2b. Microb Cell Fact. https://doi.org/10.1186/1475-2859-10-38
Gasser B, Sauer M, Maurer M, Stadlmayr G, Mattanovich D (2007) Transcriptomics-based identification of novel factors enhancing heterologous protein secretion in yeasts. Appl Environ Microbiol 73:6499–6507. https://doi.org/10.1128/AEM.01196-07
Gomar-Alba M, Alepuz P, del Olmo M, lí, (2013) Dissection of the elements of osmotic stress response transcription factor Hot1 involved in the interaction with MAPK Hog1 and in the activation of transcription. Biochim Biophys Acta 1829:1111–1125. https://doi.org/10.1016/j.bbagrm.2013.07.009
Gomez MAR, Ibba M (2020) Aminoacyl-tRNA synthetases. RNA 26:910–936. https://doi.org/10.1261/rna.071720.119
Groenewald M, Boekhout T, Neuvéglise C, Gaillardin C, Van Dijck PWM, Wyss M (2014) Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit Rev Microbiol 40:187–206. https://doi.org/10.3109/1040841X.2013.770386
Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372. https://doi.org/10.1128/mmbr.66.2.300-372.2002
Hou J, Österlund T, Liu Z, Petranovic D, Nielsen J, Osterlund T, Liu Z, Petranovic D, Nielsen J (2013) Heat shock response improves heterologous protein secretion in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:3559–3568. https://doi.org/10.1007/s00253-012-4596-9
Kobayashi Y, Iwata H, Yoshida J, Ogihara J, Kato J, Kasumi T (2015) Metabolic correlation between polyol and energy-storing carbohydrate under osmotic and oxidative stress condition in Moniliella megachiliensis. J Biosci Bioeng xx:1–6. doi: https://doi.org/10.1016/j.jbiosc.2015.02.014
Korpys-Woźniak P, Celińska E (2021) Global transcriptome profiling reveals genes responding to overproduction of a small secretory, a high cysteine- and a high glycosylation-bearing protein in Yarrowia lipolytica. Biotechnol Reports 31:e00646. https://doi.org/10.1016/j.btre.2021.e00646
Korpys-Woźniak P, Kubiak P, Białas W, Celińska E (2020) Impact of overproduced heterologous protein characteristics on physiological response in Yarrowia lipolytica steady-state-maintained continuous cultures. Appl Microbiol Biotechnol 104:9785–9800. https://doi.org/10.1007/s00253-020-10937-w
Korpys-Woźniak P, Kubiak P, Celińska E (2021) Secretory helpers for enhanced production of heterologous proteins in Yarrowia lipolytica. Biotechnol Reports 32:e00669. https://doi.org/10.1016/j.btre.2021.e00669
Kubiak M, Białas W, Celińska E (2021) Thermal treatment improves a process of crude glycerol valorization for the production of a heterologous enzyme by Yarrowia lipolytica. Biotechnol Reports 31:e00648. https://doi.org/10.1016/j.btre.2021.e00648
Kubiak M, Borkowska M, Białas W, Korpys P, Celińska E (2019) Feeding strategy impacts heterologous protein production in Yarrowia lipolytica fed-batch cultures—insight into the role of osmolarity. Yeast 36:305–318. https://doi.org/10.1002/yea.3384
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Lazar Z, Walczak E, Robak M (2011) Simultaneous production of citric acid and invertase by Yarrowia lipolytica SUC+ transformants. Bioresour Technol 102:6982–6989. https://doi.org/10.1016/j.biortech.2011.04.032
Lee J, Reiter W, Dohnal I, Gregori C, Beese-Sims S, Kuchler K, Ammerer G, Levin DE (2013) MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev 27:2590–2601. https://doi.org/10.1101/gad.229310.113
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Madzak C (2021) Yarrowia lipolytica strains and their biotechnological applications: how natural biodiversity and metabolic engineering could contribute to cell factories improvement. J Fungi 7:548. https://doi.org/10.3390/jof7070548
Madzak C (2018) Engineering Yarrowia lipolytica for use in biotechnological applications: a review of major achievements and recent innovations. Mol Biotechnol 60:621–635. https://doi.org/10.1007/s12033-018-0093-4
Mansour S, Bailly J, Delettre J, Bonnarme P (2009) A proteomic and transcriptomic view of amino acids catabolism in the yeast Yarrowia lipolytica. Proteomics 9:4714–4725. https://doi.org/10.1002/pmic.200900161
Morín M, Monteoliva L, Insenser M, Gil C, Domínguez Á (2007) Proteomic analysis reveals metabolic changes during yeast to hypha transition in Yarrowia lipolytica. J Mass Spectrom 42:1453–1462. https://doi.org/10.1002/jms.1284
Oganesyan N, Ankoudinova I, Kim SH, Kim R (2007) Effect of osmotic stress and heat shock in recombinant protein overexpression and crystallization. Protein Expr Purif 52:280–285. https://doi.org/10.1016/j.pep.2006.09.015
Park Y-K, Korpys P, Kubiak M, Celińska E, Soudier P, Trébulle P, Larroude M, Rossignol T, Nicaud J (2019) Engineering the architecture of erythritol-inducible promoters for regulated and enhanced gene expression in Yarrowia lipolytica. FEMS Yeast Res 19:1. https://doi.org/10.1093/femsyr/foy105
Pinski A, Betekhtin A, Skupien-Rabian B, Jankowska U, Jamet E, Hasterok R (2021) Changes in the cell wall proteome of leaves in response to high temperature stress in Brachypodium distachyon. Int J Mol Sci. https://doi.org/10.3390/ijms22136750
Raina M, Ibba M (2014) tRNAs as regulators of biological processes. Front Genet 5:1–14. https://doi.org/10.3389/fgene.2014.00171
Rep M, Proft M, Remize F, Tamás M, Serrano R, Thevelein JM, Hohmann S (2001) The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol 40:1067–1083. https://doi.org/10.1046/j.1365-2958.2001.02384.x
Rzechonek DA, Day AM, Quinn J, Mirończuk AM (2018) Influence of ylHog1 MAPK kinase on Yarrowia lipolytica stress response and erythritol production. Sci Rep 8:1–12. https://doi.org/10.1038/s41598-018-33168-6
Saito S, Hirao Y, Quadery AF, Xu B, Elguoshy A, Fujinaka H, Koma S, Yamamoto K, Yamamoto T (2019) The optimized workflow for sample preparation in LC-MS/MS-based urine proteomics. Methods Protoc 2:1–13. https://doi.org/10.3390/mps2020046
Schwartz CM, Hussain MS, Blenner M, Wheeldon I (2016) Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synth Biol 5:356–359. https://doi.org/10.1021/acssynbio.5b00162
Sellam A, Chaillot J, Mallick J, Tebbji F, Richard Albert J, Cook MA, Tyers M (2019) The p38/HOG stress-activated protein kinase network couples growth to division in Candida albicans. PLoS Genet 15:e1008052. https://doi.org/10.1371/journal.pgen.1008052
Sharma D, Martineau CN, Le Dall MT, Reidy M, Masison DC, Kabani M (2009) Function of SSA subfamily of Hsp70 within and across species varies widely in complementing Saccharomyces cerevisiae cell growth and prion propagation. PLoS ONE 4. https://doi.org/10.1371/journal.pone.0006644
Simpson CE, Ashe MP (2012) Adaptation to stress in yeast: to translate or not? Biochem Soc Trans 40:794–799. https://doi.org/10.1042/BST20120078
Soufi B, Kelstrup CD, Stoehr G, Fröhlich F, Walther TC, Olsen JV (2009) Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst 5:1337–1346. https://doi.org/10.1039/b902256b
Su C, Lu Y, Liu H (2013) Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Mol Biol Cell 24:385–397. https://doi.org/10.1091/mbc.E12-06-0477
Swietalski P, Hetzel F, Seitl I, Fischer L (2020) Secretion of a low and high molecular weight β-glycosidase by Yarrowia lipolytica. Microb Cell Fact 19:1–13. https://doi.org/10.1186/s12934-020-01358-5
Teige M, Scheikl E, Reiser V, Ruis H, Ammerer G (2001) Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci U S A 98:5625–5630. https://doi.org/10.1073/pnas.091610798
Theron CW, Vandermies M, Telek S, Steels S, Fickers P (2020) Comprehensive comparison of Yarrowia lipolytica and Pichia pastoris for production of Candida antarctica lipase B. Sci Rep 10:1–9. https://doi.org/10.1038/s41598-020-58683-3
Thevenieau F, Le Dall MT, Nthangeni B, Mauersberger S, Marchal R, Nicaud JM (2007) Characterization of Yarrowia lipolytica mutants affected in hydrophobic substrate utilization. Fungal Genet Biol 44:531–542. https://doi.org/10.1016/j.fgb.2006.09.001
Tomaszewska L, Rywińska A, Gladkowski W (2012) Production of erythritol and mannitol by Yarrowia lipolytica yeast in media containing glycerol. J Ind Microbiol Biotechnol 39:1333–1343. https://doi.org/10.1007/s10295-012-1145-6
Trassaert M, Vandermies M, Carly F, Denies O, Thomas S, Fickers P, Nicaud JM (2017) New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microb Cell Fact. https://doi.org/10.1186/s12934-017-0755-0
Tyanova S, Temu T, Cox J (2016a) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319. https://doi.org/10.1038/nprot.2016.136
Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016b) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. https://doi.org/10.1038/nmeth.3901
Verghese J, Abrams J, Wang Y, Morano KA (2012) Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev 76:115–158. https://doi.org/10.1128/mmbr.05018-11
Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz P-A, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus H-J, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32:223–226. https://doi.org/10.1038/nbt.2839
Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143. https://doi.org/10.1016/0003-2697(84)90782-6
Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. https://doi.org/10.1038/nmeth.1322
Wong L, Engel J, Jin E, Holdridge B, Xu P (2017) YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metab Eng Commun 5:68–77. https://doi.org/10.1016/j.meteno.2017.09.001
Yang L, Zhan X, Zheng Z, Wu J, Gao M, Lin C (2014) A novel osmotic pressure control fed-batch fermentation strategy for improvement of erythritol production by Yarrowia lipolytica from glycerol. Bioresour Technol 151:120–127. https://doi.org/10.1016/j.biortech.2013.10.031
Yang LB, Dai XM, Zheng ZY, Zhu L, Zhan XB, Lin CC (2015) Proteomic analysis of erythritol-producing Yarrowia lipolytica from glycerol in response to osmotic pressure. J Microbiol Biotechnol 25:1056–1069. https://doi.org/10.4014/jmb.1412.12026
Funding
This study was funded by the Ministry of Science and Higher Education, currently, Ministry of Education and Science, project Diamentowy Grant VII (number: DI 2017 001047).
Author information
Authors and Affiliations
Contributions
EC conceived and designed research. MKSz conducted the majority of experiments, including culturing, sample collection and analysis, and gene expression analysis. BSR and UJ conducted proteomics analysis and raw data processing. EC and MKSz analyzed data and did bioinformatics. EC and MKSz wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kubiak-Szymendera, M., Skupien-Rabian, B., Jankowska, U. et al. Hyperosmolarity adversely impacts recombinant protein synthesis by Yarrowia lipolytica—molecular background revealed by quantitative proteomics. Appl Microbiol Biotechnol 106, 349–367 (2022). https://doi.org/10.1007/s00253-021-11731-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11731-y