Abstract
Acetic acid bacteria (AAB) are valuable biocatalysts for which there is growing interest in understanding their basics including physiology and biochemistry. This is accompanied by growing demands for metabolic engineering of AAB to take advantage of their properties and to improve their biomanufacturing efficiencies. Controlled expression of target genes is key to fundamental and applied microbiological research. In order to get an overview of expression systems and their applications in AAB, we carried out a comprehensive literature search using the Web of Science Core Collection database. The Acetobacteraceae family currently comprises 49 genera. We found overall 6097 publications related to one or more AAB genera since 1973, when the first successful recombinant DNA experiments in Escherichia coli have been published. The use of plasmids in AAB began in 1985 and till today was reported for only nine out of the 49 AAB genera currently described. We found at least five major expression plasmid lineages and a multitude of further expression plasmids, almost all enabling only constitutive target gene expression. Only recently, two regulatable expression systems became available for AAB, an N-acyl homoserine lactone (AHL)-inducible system for Komagataeibacter rhaeticus and an l-arabinose-inducible system for Gluconobacter oxydans. Thus, after 35 years of constitutive target gene expression in AAB, we now have the first regulatable expression systems for AAB in hand and further regulatable expression systems for AAB can be expected.
Key points
• Literature search revealed developments and usage of expression systems in AAB.
• Only recently 2 regulatable plasmid systems became available for only 2 AAB genera.
• Further regulatable expression systems for AAB are in sight.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acetic acid bacteria (AAB) are a group of obligately aerobic Gram-negative bacteria that exhibit a unique form of metabolism by which they typically partially oxidize a variety of substrates such as sugars or ethanol by membrane-bound dehydrogenases (mDHs) and produce acetic acid. AAB are already used for a long time and they are very important for the production of foods and beverages such as vinegar, kombucha, kefir, and other products. Several reviews on AAB are already available that provide overviews, discussions and outlooks on issues related to their taxonomy, physiology and biochemistry, fermentation (foods and beverages), acetic acid resistance, production of exopolysaccharides including bacterial cellulose, pyrroloquinoline quinone (PQQ)-dependent dehydrogenases, oxidation of carbon sources and alcohols, nitrogen fixation, metabolic engineering, biotechnological and industrial applications, as well as their use as microbial biosensors (see, for example, Cleenwerck and De Vos 2008; De Roos and De Vuyst 2018; Gao et al. 2020b; Gomes et al. 2018; Gullo et al. 2018; Kolesovs and Semjonovs 2020; La China et al. 2018; Laureys et al. 2020; Lynch et al. 2019; Mamlouk and Gullo 2013; Matsutani and Yakushi 2018; Trcek et al. 2015; Wang et al. 2015; Zhang et al. 2020). No review is currently available that provides an overview of the expression systems developed, tested and used for target gene expression and their use cases in AAB.
Generally, the expression of target genes to produce proteins in bacterial cell culture for various purposes is a standard method in basic research and biotechnological applications. For constitutive and inducible or regulatable expression, numerous plasmids have already been developed and established in many bacteria, which have been summarized and reviewed in the past for bacteria other than AAB (see, for example, Chen 2012; Connell 2001; Dilworth et al. 2018; Evans and Mizrahi 2015; Forstner et al. 2007; Gruber et al. 2015; Parachin et al. 2012; Schnappinger and Ehrt 2014; Terpe 2006; Valero 2012). In AAB, for a long time, only constitutive target gene expression and in a very few studies only weakly regulatable target gene expression was achieved since, for example, the transfer of heterologous regulatable systems such as the well-known classical examples of the TetR-, AraC-, and LacI-dependent systems and their application was not really successful yet in AAB due to high (leaky) expression already in the absence of the respective inducer (Florea et al. 2016a; Teh et al. 2019). Only two regulatable expression systems, an N-acyl homoserine lactone-inducible luxR-Plux system for Komagataeibacter rhaeticus and an l-arabinose-inducible araC-ParaBAD system for Gluconobacter oxydans with up to 480-fold induction have recently been reported for the two AAB species (Florea et al. 2016a; Fricke et al. 2020). To provide a comprehensive overview of expression systems, their latest developments and the use cases in AAB we have searched and evaluated the literature for relevant AAB studies.
Literature search and updates in the systematics of AAB genera
We carried out a keyword-based literature search using the Web of Science Core Collection (WoSCC) database. Currently, for the Acetobacteraceae family there are 49 genera listed in the taxonomy browser of the NCBI website. We assumed that a publication potentially relevant for finding expression plasmids in AAB should contain at least the name of one of these AAB genera in the title, abstract, or keywords, respectively (for Stella we used the two full names Stella humosa and Stella vacuolata). In this first literature filtering step, the single AAB genus word-based searches in the WoSCC database revealed that the top 3 AAB genera with by far the most publications were Acetobacter (3,774), Gluconobacter (1,446) and Glucon(o)acetobacter (1,194), respectively. These top 3 genera were followed by Acidiphilium (345), Komagataeibacter (229), Roseomonas (177), Asaia (175), Acidocella (62) and Acidomonas (33). As outlined below, some genera names were introduced only recently and many of the included species had a different genus name before. Together, these 9 AAB genera already represent 97.6% (6,288) of all (6,440) non-redundant publications somehow related to one or more out of the 49 AAB genera. We then selected the 6097 publications from the year 2020 backward to 1973, the year of the first published successful DNA cloning experiments in Escherichia coli (Cohen et al. 1973). Using EndNote, for 3397 publications (55.7%), we could directly get access to the full text (PDF) due to Open Access or journal access via our institutions’ central library. To narrow down the AAB-related publication list in a second step, we filtered title, abstract, keywords, and, if available, the full text (PDF) on the one hand by checking for the presence of the text strings plasmid, vector, host, induction, inducer, activation, activator, repression, repressor, expression, promoter, regulation or terminator, respectively. On the other hand, we also checked the publications by manual inspection of title, abstract and, if available, full-text PDF in regard to expression systems, the genetic work and the case studies. We finally ended up with a list of 243 publications from 1985 onward representing studies in which plasmids have been created, tested or used in 9 AAB genera. We probably missed a number of studies reporting plasmids for expression in AAB due to missing full-text information.
It should be mentioned that the taxonomy of the AAB has gone through some updates including the creation of new AAB genera and renaming of AAB species already described in the literature. In 1989 the genus Acidomonas was proposed to include a group of acidophilic, facultatively methylotrophic bacteria (Urakami et al. 1989). These microorganisms are Gram-negative, non-spore forming, non-motile, and rod-shaped and grow at pH 2.0 to 5.5. These characteristics are unique among the methanol-utilizing bacteria and the typical strain Acetobacter methanolicus can be distinguished from the type and representative strains of Acetobacter, Gluconobacter, and Acidiphilium, resulting in the renaming of Acetobacter methanolicus to Acidomonas methanolica. In 1997 a genus concept of AAB was proposed classifying Acetobacter, Gluconoacetobacter, Acidomonas, and Gluconobacter based on partial 16S rRNA sequences (Yamada et al. 1997). In 1998 Gluconoacetobacter was changed to Gluconacetobacter (International Committee on Systematic Bacteriology). As a consequence of these changes and updates, Acetobacter diazotrophicus, A. europaeus, A. hansenii, A. liquefaciens, and A. xylinum (alias A. xylinus) were included in the genus Gluconacetobacter with the respective changes in nomenclature: Gluconacetobacter diazotrophicus, Ga. europaeus, Ga. hansenii, Ga. liquefaciens, and Ga. xylinus, respectively. Furthermore, in the following years, a phylogenetic duality in the new genus Gluconacetobacter was found. The Ga. liquefaciens group and the Ga. xylinus group could be phylogenetically, phenotypically and ecologically distinguished from each other at the generic level. This resulted in the creation of the new AAB genus Komagataeibacter based on 16S rRNA gene sequences and the transfer of several new combinations including Ga. xylinus (formerly A. xylinus alias A. xylinum) to Komagataeibacter xylinus on the basis of taxonomic characteristics (Yamada 2014; Yamada et al. 2012). Taking these AAB genera updates and renaming of AAB species into account for the publications before 1989, 1998, and 2014 (and after), we found in our final list 101 publications related to Gluconobacter, 61 to Komagataeibacter, 41 to Acetobacter, 20 to Gluconacetobacter, 8 to Acidiphilium, 6 to Acidomonas, 4 to Asaia, 1 to Kozakia, and 1 to Roseomonas, respectively. Among these 243 publications, in 59 studies genomic allele replacements and transposon- or plasmid-based gene inactivation screenings requiring the functional expression of transposases or resistance genes as well as homologous recombination were carried out using a few strategies without further recombinant target gene expression. These studies were excluded from our review which removed the genera Kozakia and Roseomonas from our list. The remaining 184 publications were organized according to the expression plasmids used, the case studies or their timelines, and are presented for the seven remaining AAB genera according to the number of publications (Fig. 1).
Target gene expression in Gluconobacter
Most studies reporting on the construction and usage of expression plasmids in AAB were found for the genus Gluconobacter (81). Initially, in 11 studies, several plasmids were constructed and tested before a major expression plasmid family was established in Gluconobacter in 2006.
Plasmid diversity before establishing the major expression plasmid lineage
The first study reporting an E. coli / Gluconobacter shuttle vector was published in 1985 by Teruhiko Beppu and co-workers, who in parallel were also working on shuttle vectors for Acetobacter (see Acetobacter section). This first shuttle vector for Gluconobacter was constructed by ligation of cryptic plasmid pMV201 (2.5 kb) found in G. suboxydans IFO 3130 with the E. coli plasmid pACYC177 (3.9 kb) carrying the p15A ori. The resulting chimeric plasmid pMG101 (6.2 kb) carries an ampicillin (Amp) resistance gene and replicates in G. suboxydans as well as in E. coli (Fukaya et al. 1985b). Later, conjugal transfer of a series of plasmids of the incompatibility group P (RP4, RP1::Tn951, pRK290 and derivatives thereof) and Q (pKT230/231) was studied. A gentamycin (Gm) resistance-encoding pRK290 derivative (20 kb, minimal replicon derivative of RK2) was constructed and suggested to offer considerable potential as a versatile gene delivery system for Gluconobacter (Condon et al. 1991). The promoter of the Gm resistance gene is functional since pRK290::Gm increased the minimal inhibitory concentration for gentamycin from 80 μg/mL to more than 2 mg/mL. With plasmid RP1::Tn951 carrying the lactose transposon, heterologous gene expression and regulation was tested in Gluconobacter. Tn951 contains IPTG-inducible and cAMP receptor protein (CRP)-dependent genes homologous to the E. coli lacI, lacZ, and lacY genes. The Tn951-based β-galactosidase activity was found to be induced 4-fold to 6-fold by IPTG in Gluconobacter, which suggested the synthesis of a functional LacI repressor in Gluconobacter, yet the repression was leaky in the absence of IPTG and the induced LacZ activity less than 5% compared to fully induced levels in E. coli. Since in E. coli the lac operon displays reversible cAMP-dependent catabolite repression with glucose, cAMP was exogenously added (100 μM) to the Gluconobacter culture. cAMP did not result in any detectable increase in LacZ activity, suggesting that a catabolite repression-like phenomenon was unlikely to be the cause of the poor LacZ activity in Gluconobacter if the assumption holds true that cAMP can be transported into the cell at levels sufficient for physiological effects to be observed. With the genome sequences available today we know that a CRP gene appears to be absent in Gluconobacter. The low levels of LacZ activity could be explained by inefficient recognition of the heterologous Tn951 promoter in Gluconobacter (Condon et al. 1991).
Generally, RK2-based vectors such as the broad host range vector pRK293 (21.4 kb) can be transferred to G. oxydans by bacterial mating, yet efforts to use this cloning vehicle were unsuccessful, since the transfer of pRK293 occurred only with very low efficiency and with pRK293 containing a 4.7 kb insert of interest no clones could be obtained (Cleton-Jansen et al. 1991). In another attempt to construct and test a suitable shuttle vector, the small cryptic plasmid pGY1 (2.7 kb) also detected in G. suboxydans IFO 3130 was cleaved and ligated with cleaved pUC18 (Takeda and Shimizu 1992). The resulting chimeric vector pGEA1 (5.4 kb) carries ampicillin resistance and was used to express the cytochrome c-553 gene of G. suboxydans IFO 12528 from its native promoter region in G. suboxydans IFO 3254, which oxidizes ethanol poorly because of a deficiency of the second subunit of the membrane-bound alcohol dehydrogenase. Transformants of IFO 3254 exhibited ethanol oxidation activity and increased dehydrogenase activities for d-glucose, d-sorbitol and glycerol. Furthermore, azide insensitivity of the respiratory chain was restored, which indicated that cytochrome c-553 is in the pathway of the azide-insensitive respiratory chain bypass of G. suboxydans (Takeda and Shimizu 1992; Takeda et al. 1992). While in these early Gluconobacter studies chemically competent cells or conjugation were used to transfer plasmids, electroporation was also established for Gluconobacter and enabled transformation frequencies of up to 105 transformants /μg of DNA (Creaven et al. 1994). In the following, conjugation and electroporation were used for Gluconobacter almost equally often.
In a further attempt to construct, test, and establish a suitable shuttle vector, the chimeric plasmid pGE1 (11.9 kb) was constructed from the endogenous cryptic plasmid pGO3293S (9.9 kb) found in G. oxydans IFO 3293 and plasmid pSUP301 (5 kb) from E. coli (Shinjoh and Hoshino 1995). The plasmid pGO3293S encodes enzyme activities that converts l-sorbose to 2-keto-l-gulonic acid, the non-lactonized precursor of vitamin C. Plasmid pSUP301 contains pACYC177 introduced above. The plasmid pGE1 could be transferred into G. oxydans IFO 3293 with a high frequency (10-1 transconjugants per recipient) by conjugal transfer, maintained very stably without antibiotic selection, and did not inhibit the growth or 2-keto-l-gulonic acid productivity of producer strains derived from G. oxydans IFO 3293. pGE1 could also be shortened to a 9.8-kb plasmid termed pGE2. pGE1 could be introduced in 6 Gluconobacter and 4 Acetobacter strains out of 6 and 28 strains tested. Thus, it showed a broader host-range than the shuttle vector pMG101 introduced above, which showed a limited host-range even in Gluconobacter. pGE1 was considerably stable in the transconjugants after two cycles of cultivation without kanamycin, except in G. frateurii IFO 3271. The usefulness of pGE1 as an expression vector was confirmed by subcloning the membrane-bound l-sorbosone dehydrogenase (SNDH) gene of A. liquefaciens IFO 12258 and its expression in G. oxydans IFO 3293 derivatives. Using resting cells, pGE1 derivatives with the SNDH gene, however, did not lead to a higher 2-keto-l-gulonic acid production than the pVK102 derivative with the SNDH gene (Shinjoh and Hoshino 1995). Already beforehand, plasmid pVK102 (23 kb), a mobilizable broad-host-range cosmid vector constructed from pRK290 and pHK17 via the intermediate pVK100 originally used in Agrobacterium and providing kanamycin and tetracycline resistance, was used to construct a genomic library of A. liquefaciens IFO 12258 for a SNDH activity screening in a G. oxydans IFO 3293 mutant that accumulates l-sorbosone in the presence of l-sorbose (Shinjoh et al. 1995). Plasmid pVK100 was used in a check of the biological functions of the polyol dehydrogenase genes sldAB to complement mutants of G. oxydans DSM 4025 disrupted in the sldAB genes by expressing sldA, sldB, or both genes from G. oxydans promoter PA (Shinjoh et al. 2002).
In another attempt to create a shuttle vector, the endogenous plasmid pF4 (4.4 kb) extracted from G. oxydans T-100 was ligated with plasmid pHSG298 (2.7 kb) carrying pBR322 ori and the resulting chimeric plasmid pFG15A (7.3 kb) was confirmed to be also stable in G. oxydans G624 across several passages in a broth without kanamycin and had no effects on the l-sorbose production by the host (Saito et al. 1997). Plasmid pFG15A was used to express l-sorbose and l-sorbosone dehydrogenase genes of G. oxydans T-100 from their native promoter regions. G. oxydans G624, which is unable to produce 2-keto-l-gulonic acid endogenously, cultivated in the presence of d-sorbitol and with the expression plasmid reached a 2-keto-l-gulonic acid titer 2.3-fold higher than the titer obtained by G. oxydans T-100, yet the yield was still insufficient (Saito et al. 1997). Therefore, to improve the expression of the dehydrogenase genes, the native promoter region was replaced with that of E. coli PtufB1, Plac, or PL, which all were active. In combination with chemical mutagenesis of the strain, the 2-keto-l-gulonic acid production level was further increased 3-fold (Saito et al. 1998).
In another study, the broad-host-range vector pSUP104, which includes the mobilization and replication functions of the IncQ plasmid RSF1010 and was originally tested in Rhizobium, Agrobacterium, and Pseudomonas species, respectively, was used in Gluconobacter to complement a Tn5-based pqqE-deficient mutant by expressing a genomic library fragment containing the pqqE gene of the PQQ coenzyme biosynthesis gene cluster pqqBCDE of G. oxydans ATCC 9937 (Felder et al. 2000). RSF1010 was used otherwise in some Acidomonas studies and in an ori test using the pSEVA tool kit in Komagataeibacter (see sections below).
Another expression plasmid used in Gluconobacter is the shuttle vector pSA19 already developed and introduced for use in Komagataeibacter in 1994 (see Komagataeibacter section). The feasibility of using pSA19 was also demonstrated for Gluconobacter by expressing the xylitol dehydrogenase (XDH) gene of Morganella morganii from the promoter Plac (Tonouchi et al. 2003). In the recombinant G. oxydans ATCC 621 strain, the XDH activity was increased 4-fold. To develop an improved production process for the alternative sweetener xylitol, also the endogenous XDH gene from G. oxydans ATCC 621 was cloned with and without its native promoter region into pSA19 (Sugiyama et al. 2003). Expression from Plac alone increased the XDH activity 5-fold in the recombinant G. oxydans strain, while XDH activity was 11-fold higher when the gene was expressed from both Plac and its native promoter. The latter construct increased the xylitol titer obtained by conversion of d-arabitol to xylulose with arabitol dehydrogenase followed by reduction to xylitol 2-fold within 48 h when ethanol was added. Later pSA19 was used to express the sboA and sboR genes both located upstream of the FAD-dependent d-sorbitol dehydrogenase (SLDH) genes sldSLC in G. frateurii THD32 to analyze SboA enzyme activity and the role of the putative transcriptional regulator SboR in the regulation of sldSLC expression (Soemphol et al. 2007). The SboA enzyme showed NADPH-dependent l-sorbose reductase activity. The predicted transcriptional regulator SboR was suggested to be a repressor of sboA expression while another regulator was assumed to be required for the induction of sldSLC by d-sorbitol or l-sorbose. When sboR was disrupted in G. frateurii, the SboA activity was increased 1.5-fold and 2-fold when grown on d-sorbitol and l-sorbose, respectively. Since the basal expression in the parental strain was already relatively high according to the activities in the cytosolic fraction (0.6 to 0.9 U/mg) the SboR-repressed promoter of the sboRA operon seems not suitable to be used for a regulatable expression system in Gluconobacter.
In parallel to the test and usage of pSA19 in Gluconobacter, another shuttle plasmid was created and tested. The cryptic endogenous plasmid pAG5 (5.6 kb) found in G. oxydans IFO 3171 was ligated with pUC18 and the chimeric vector was termed pSG8 (Tonouchi et al. 2003). Additionally, the 2 kb smaller plasmid pSG6 was created by EcoRI digestion of pSG8 and self-ligation of the 3.6 kb fragment. The copy numbers of pSG8/6 were estimated to be 10 per genome and both were found to be stable in G. oxydans ATCC 621 after 10 days of repeated cultivation (50 generations) in the absence of ampicillin. Plasmid pSG6 was used for expression of the endogenous cyanide-insensitive quinol oxidase genes cioAB in the wild type G. suboxydans IFO 12528 (Mogi et al. 2009). Additional plasmid-based expression was required for the biochemical characterization of the membrane-bound enzyme, as it was difficult without due to the low native expression level and the enzyme instability in detergent solution.
pBBR1MCS derivatives—the major plasmid family used in Gluconobacter
Besides testing the various plasmids described above for recombinant expression, in 2002, the plasmid pBBR122 was used to test whether lactose metabolism could be established in G. oxydans by cloning and expressing the E. coli lacZY genes from Plac in five G. oxydans strains and one Ga. liquefaciens strain (Mostafa et al. 2002). In three G. oxydans strains the LacZ activities were similarly high as the LacZ activity in induced E. coli XL1-Blue cells, while on other G. oxydans strains and in Ga. liquefaciens the LacZ activities were much lower. Albeit for unknown reasons only a few transformants of G. oxydans were able to grow on lactose, these results demonstrated that active β-galactosidase can be produced in G. oxydans at levels much higher than the LacZ activities obtained with Tn951 mentioned above. This also suggested that the Tn951 promoter is very weak in Gluconobacter which could be useful when very low target gene expression levels are required.
The plasmid pBBR122 is based on the very small endogenous plasmid pBBR1 (2.6 kb) isolated from Bordetella bronchiseptica (Antoine and Locht 1992). pBBR122 was commercialized by MoBiTec and exhibits a broad-host-range compatible with IncP, IncQ, and IncW group plasmids as well as with ColE1- and p15A-based replicons. The pBBR1 derivative pBBR1MCS carrying pBBR1 ori, a MCS and a chloramphenicol resistance cassette was used to create four new derivatives each with a different antibiotic resistance cassette: pBBR1MCS-2 confers kanamycin resistance, pBBR1MCS-3 tetracycline resistance, pBBR1MCS-4 ampicillin resistance, and pBBR1MCS-5 gentamicin resistance (Kovach et al. 1995; Kovach et al. 1994). The family members exhibit several advantages in that they are relatively small (<5.3 kb), possess an extended MCS, allow direct selection of recombinant plasmids in E. coli via disruption of the LacZα peptide, and are mobilizable when the RK2 transfer functions are provided in trans, yet can also be introduced by electroporation.
Since 2006, the pBBR1MCS-based plasmids have been used in most Gluconobacter studies reporting recombinant gene expression; thus, it is the major expression plasmid lineage used in Gluconobacter and it also exhibits the most use cases in AAB in general (Table 1). While pBBR1MCS-3 providing tetracycline resistance was apparently not used in Gluconobacter, pBBR1MCS-5 has been used in 27 studies, pBBR1MCS-2 in 25 studies, and pBBR1MCS-4 in 8 studies. In 51 cases Gluconobacter genes, in 5 cases genes from other AAB genera, and in 13 cases genes from bacteria other than AAB, were expressed from Plac or selected promoters (Table 2). In a few studies, fusion proteins with signal peptides for export or for surface display and a tagged protein for one-step membrane protein purification from Gluconobacter cells were produced.
Other expression plasmids recently used in Gluconobacter
In parallel to the use of the very successful pBBR1MCS-based plasmid family in Gluconobacter, further plasmids have been constructed based on the two homologous cryptic plasmids pGOX3 (14.5 kb) found in two G. oxydans strains and pUC18 or pUC19 carrying pBR322 ori. The chimeric plasmids are selectable by conferring ampicillin resistance, are compatible with the pBBR1MCS family and double selectable except with pBBR1MCS-4 also carrying ampicillin resistance. In the first case, the par and rep loci of pGOX3 from G. oxydans DSM 2003 were amplified as a 2.3-kb fragment and cloned into pUC18, resulting in the 5 kb shuttle vector pZL1 (Zhang et al. 2010). Plasmid pZL1 was found to replicate in both E. coli and G. oxydans DSM 2003 and was maintained for 144 h during serial subcultures in the absence of selective pressure in 80% of the DSM 2003 cells, while pUC18 was almost completely lost already after 48 h. The relative plasmid copy number of pZL1 in DSM 2003 was found to be 13 times higher than that of the endogenous pGOX3. The capability of pZL1 to express heterologous genes in DSM 2003 was shown by using the fluorescence reporter gene wasabi. When expressed in DSM 2003 from the strong promoter PtufB, the fluorescence signal with the reporter plasmid pZL1-tufB-wasabi was almost as high as with pBBR1MCS-5-tufB-wasabi.
A comparable assembly process as for pZL1 was used to construct the shuttle vector pGUC based on the par and rep loci of pGOX3 from G. oxydans 621H and pUC18 (Gao et al. 2014). Plasmid pGUC and again the strong promoter PtufB were used to express and test different combinations of five l-sorbose dehydrogenase (SDH) genes and two l-sorbosone dehydrogenase (SNDH) genes from Ketogulonicigenium vulgare in G. oxydans WSH-003, an industrial strain used for the conversion of d-sorbitol to l-sorbose. As the production of the vitamin C precursor 2-keto-l-gulonic acid from d-sorbitol involves three sequential oxidation reactions, the spatial proximity of the corresponding dehydrogenases appeared to be useful to enhance the production. Therefore, with a series of linker peptides, SDH-SNDH fusion enzymes were tested and for a selected SDH-SNDH fusion the pqqA gene alone or the pqqABCDE gene cluster for PQQ biosynthesis was additionally included on the pGUC plasmid. Additional expression of the PQQ biosynthesis gene(s) enhanced cell growth and with the stepwise metabolic engineering of G. oxydans, the final 2-keto-l-gulonic acid titer was improved 8-fold (39 g/L) compared to that obtained with the independent expression of the dehydrogenase genes (Gao et al. 2014). In further work, the adaptor protein SH3 gene sequence was fused via a linker sequence to the SDH gene of the SDH-linker-SNDH construct and co-expressed with the gene of the small trimeric protein CutA from Pyrococcus horikoshii fused to the SH3lig gene sequence, all present on pGUC and each expressed from PtufB (Gao et al. 2020c). The adaptor protein SH3 as docking protein and its ligand SH3lig as docking station peptide improved the chemical structure stability of fused SDH-SNDH complexes surrounding the trimeric CutA protein. The recombinant strain G. oxydans WSH-003 with the pGUC-based expression plasmid produced 40 g/L of 2-keto-l-gulonic acid after 168 h. Additionally, co-expression of the pqqABCDE operon from PtufB on pGUC increased the 2-keto-l-gulonic acid titer to 43 g/L, demonstrating an efficient conversion of d-sorbitol to 2-keto-l-gulonic acid with a single strain (Gao et al. 2020c).
In another study, the amplified par-rep fragment of pGOX3 from G. oxydans 621H was ligated with pUC19 and shuttle vector pUCpr was obtained (Yuan et al. 2016a). In this work two compatible plasmids, pBBR1MCS-5 and pUCpr have been applied simultaneously in the same strain for target gene expression. Plasmid pBBR1MCS-5 was used to express the membrane-bound PQQ-dependent sorbitol dehydrogenase genes sldAB from the strong promoter PGOX0169 in different combination with pUCpr derivatives carrying either the PQQ biosynthesis genes pqqABCDE or pqqABCDE-tldD under control of PGOX0169 alone, or each in combination with the cytochrome bo3 oxidase genes cyoBACD expressed from a separate copy of the PGOX0169 promoter. In fed-batch cultivation with pH and dissolved oxygen tension control, 5-keto-d-gluconate production could be significantly enhanced with the combinatorial metabolic engineering strategy to 162 g/L based on the two-plasmid-system in the recombinant industrial G. oxydans strain ZJU2 (Yuan et al. 2016a).
Plasmid-based promoter analysis in Gluconobacter
Besides the expression of target genes in screening, complementation, functional, or metabolic engineering studies, plasmids have also been used in Gluconobacter to analyze and compare relative promoter strength using selected reporter genes. The promoter probe vector pCM130 is a derivative of the Plac expression plasmid pCM62 introduced below in the Acetobacter section and carries the reporter gene xylE encoding catechol 2,3-dioxygenase from Pseudomonas in place of Plac. It was used to compare the strength of the promoter PpqqA and a potential intrinsic promoter PpqqBCDE of the pqqABCDE operon from G. oxydans 621H (Hölscher and Görisch 2006). With both d-mannitol or d-gluconate as a growth substrate, the reporter activities of PpqqBCDE were below the activity found for the pCM130 empty vector, while with PpqqA the reporter activities were at least 3.5-fold higher than with the empty vector. Therefore, the reporter assay results suggested that PpqqA represents the only promoter of the pqqABCDE operon in G. oxydans 621H, a situation similar to that found for other PQQ-synthesizing bacteria.
With the aim to provide effective Plac-independent expression vectors for gene expression in G. oxydans, constitutive promoters were selected to determine their relative strength using pBBR1MCS-2 and the β-d-glucuronidase gene uidA from E. coli as a reporter (Kallnik et al. 2010). The promoters of GOX0264 and GOX0452 encoding the ribosomal proteins L35 and L13, respectively, were found to be strong (PGOX0264) and moderate (PGOX0452), while the intrinsic promoter Plac of pBBR1MCS-2 appeared rather weak in G. oxydans 621H. In another approach to identify strong promoters, chromosomal DNA of G. oxydans DSM 2003 was randomly digested and DNA fragments were inserted into pBBR1MCS-5-gfp carrying the promoterless green fluorescent protein gene as a reporter (Shi et al. 2014). After screening the GFP fluorescence signals of 710 transformants, the one with the highest fluorescence intensity was selected and a 261-bp DNA fragment was obtained that exhibited promoter activity and was homologous to the G. oxydans 621H intergenic region between GOX0168 encoding an NAD-dependent DNA ligase and GOX0169 encoding a hypothetical protein. The promoter region was termed gHp0169 and PgHp0169 activity was determined to be almost 2-fold stronger (2.47 U/mg) than PtufB from G. oxydans (1.39 U/mg) when assayed with pBBR1MCS-5 and the ndh gene encoding the type II NADH dehydrogenase as a reporter.
Since strong promoters are one of the essential factors that can improve strain performance by overexpression of specific genes, a pipeline for screening strong promoters by proteomics analysis and promoter assays was also established (Hu et al. 2015). Using this approach, the strong promoter PB932_2000 was identified in G. oxydans WSH-003. As assessed by analysis of pBBR1MCS-2-based egfp expression and qRT-PCR analysis of egfp mRNA, the strength of PB932_2000 in G. oxydans WSH-003 was at least 2-fold higher than that of PtufB from G. oxydans and at least 4-fold higher than that of PtufB from E. coli. In a further study the aforementioned G. oxydans promoters PtufB, PGOX0169, PGOX0264, and PGOX0452 were assayed for their relative strength in G. oxydans 621H using pBBR1MCS-5 and gfp as reporter gene (Yuan et al. 2016a). In this assay, PGOX0169 was found to be the strongest one, followed by PGOX0264, PtufB, and PGOX0452, respectively.
In contrast to the previous studies searching mainly for strongest promoters, another study aimed to develop a new vector for successful expression of genes encoding membrane-bound dehydrogenases in G. oxydans, which typically requires intermediate or even low expression levels to obtain functional enzymes (Mientus et al. 2017). In this study, the strength and the regulation of the promoters of the alcohol dehydrogenase gene (Padh) and the inositol dehydrogenase gene (Pidh) were analyzed using pBBR1MCS-5-based pJV17 derivatives carrying the E. coli lacZ reporter gene. According to the β-galactosidase assays, both promoters were practically not active in E. coli, while they showed good activity in G. oxydans grown on d-mannitol, d-sorbitol or d-glucose as carbon source. Padh led to similar LacZ activities with all three substrates and was generally more active than Pidh, which led to weak LacZ activity on d-mannitol and appeared to be repressed 100-fold in the presence of d-glucose when compared to d-sorbitol.
Chromosomal integrations and intergenic regions suitable for target gene expression
Besides plasmid-based expression, chromosomally integrated expression cassettes using either the native promoter or selected endogenous or heterologous promoters have also been applied for target gene expression in Gluconobacter. In an early study analyzing the maltose-oxidizing ability of a G. oxydans strain, the effect of a single amino acid substitution on the substrate specificity of the PQQ-dependent glucose dehydrogenase (mGDH) was tested by chromosomal insertion due to the lack of a suitable expression plasmid for Gluconobacter at that time (Cleton-Jansen et al. 1991). A G. oxydans strain with an endogenous mGDH not capable of oxidizing maltose was transformed with the non-replicable pGP173 plasmid backbone, which carried the mGDH gene of the maltose-oxidizing strain under control of its native promoter region and a kanamycin resistance cassette. Recombinant cells formed by homologous recombination were selected by growth on maltose. Further analysis revealed that the exchange H787N broadened the substrate spectrum of the mGDH and enabled the oxidation of the disaccharide maltose.
Later, chromosomal integrations for target gene expression have been used in combinatorial metabolic engineering studies for which suitable integration sites had to be selected. For example, to increase the biomass yield of G. oxydans 621H on glucose, of which 90% is typically oxidized already in the periplasm to gluconate and 2-ketogluconate accumulating unused in the medium, the carbon flux into the central carbon metabolism needs to be increased and additionally, the incomplete tricarboxylic acid (TCA) cycle of G. oxydans 621H needs to be completed by a succinate dehydrogenase and a succinyl-CoA synthetase, since these two TCA cycle enzymes are absent. These requirements were addressed by combining the chromosomal insertion of several heterologous genes and deletion of the two glucose dehydrogenase genes to eliminate the periplasmic and cytosolic oxidation of glucose to gluconate (Kiefler et al. 2017). In detail, the cytoplasmic NADP-dependent glucose dehydrogenase gene gdhS was replaced by the succinate dehydrogenase genes sdhCDAB and the flavinylation factor gene sdhE from A. pasteurianus with the sdhCDAB operon under the control of strong PGOX0264 and sdhE under the control of its native promoter. The membrane-bound PQQ-dependent glucose dehydrogenase gene gdhM was replaced by the succinyl-CoA synthetase genes sucCD from Ga. diazotrophicus again using the strong promoter PGOX0264. Furthermore, the pyruvate decarboxylase gene pdc was replaced by a second ndh gene for a type II NADH dehydrogenase with its native promoter from G. oxydans DSM 3504 to eliminate acetate formation from pyruvate and to increase the overall NADH oxidation capacity. Together, the biomass yield of the engineered plasmid-free G. oxydans strain IK003.1 was increased by 60% and the strain was stable for more than 70 generations, making it very interesting as a host for oxidative biotransformations and further metabolic engineering approaches (Kiefler et al. 2017; Kranz et al. 2017).
Generally, for successful expression of chromosomally integrated recombinant genes, suitable intergenic regions need to be identified. Consequently, G. oxydans IK003.1 was used to test different intergenic regions (IGRs) selected on the basis of RNAseq data for their suitability to integrate and express the fructose dehydrogenase (FDH) genes fdhSCL from G. japonicus under the control of the strong promoter PGOX0264 for the oxidative biotransformation of fructose to 5-ketofructose (5-KF) (Battling et al. 2020; Kranz et al. 2018). 5-KF is a promising non-nutritive natural sweetener for which high-titer production was achieved already before in plasmid-based approaches (Kawai et al. 2013; Siemen et al. 2018). In the chromosome-based approach, four IGRs have been identified in the G. oxydans 621H genome suitable for expression of heterologous genes. That is the IGR of the genes GOX0013-GOX0014, GOX0028-GOX0089; GOX0038-GOX0039 and GOX2095-GOX2096, respectively, all located close to the genomic ori to take advantage of the increased average DNA copy number during chromosome replication in the exponential growth phase. Finding more than one suitable integration site allowed independent double-integration of the fdhSCL operon associated with a strong increase in the 5-KF production rate compared to the single integration strains. Therefore, more integrated copies of the fdhSCL operon are expected to further increase the 5-KF production rate by plasmid-free strains. Additionally, the use of a promoter stronger than PGOX0264 could enhance fdhSCL expression and thereby 5-KF production. Besides fructose as substrate, 5-KF could also be produced from the cost-efficient and renewable feedstock sucrose when applying a sucrose-hydrolyzing enzyme and FDH (Hoffmann et al. 2020). The fdhSCL genes were disruptively integrated into the mGDH gene locus under the control of the strong promoter PGOX0264 and the invertase Inv1417 gene was expressed under the control of the same promoter on a pBBR1MCS-2-based plasmid. The average yield of 5-KF formation from sucrose was 90 mM 5-KF, which is 84% in relation to the fructose released from 105 mM sucrose almost completely hydrolyzed. The invertase contains a Tat-signal peptide for secretion into the periplasm, but only 40% of the total invertase activity was found in the periplasm and 60% in the cytoplasm. This suggested that plasmid-based expression of the Inv1417 gene using the strong promoter PGOX0264 possibly caused an overload of the Tat export machinery. Chromosomal integration of the invertase gene into one of the suitable G. oxydans IGRs mentioned above might solve this limitation.
The same principle of combining gene deletion to eliminate undesired enzyme activity and inserting an expression cassette into the disrupted gene locus was applied for biotransformation of glucose to 2-keto-d-gluconic acid via intermediate d-gluconic acid by recombinant G. japonicus. The gene for a subunit of gluconate-5-dehydrogenase producing the unwanted by-product 5-keto-d-gluconic acid from d-gluconic acid was deleted and in this locus a cassette for the expression of a gluconate-2-dehydrogenase gene was inserted to increase 2-keto-d-gluconic acid formation from d-gluconic acid produced from glucose (Zeng et al. 2019).
l-Arabinose-induced AraC-dependent regulatable expression
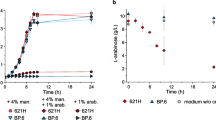
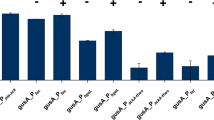
Until now, an l-arabinose-inducible araC-ParaBAD system on the pBBR1MCS-5 backbone is the only regulatable system available for Gluconobacter (Fricke et al. 2020). The mechanism of transcriptional gene regulation by AraC is different compared to those of the pure repressor proteins TetR and LacI, which just dissociate from their operator DNA when their respective inducer is bound to the protein. In E. coli AraC does not only repress ParaBAD by looping the DNA when bound to specific target sequences in the absence of l-arabinose, but also is essential for activation of ParaBAD (Fig. 2a). In the presence of l-arabinose, a modified binding of AraC to target sequences causes unlooping of the promoter DNA and stimulation of both the binding of RNA polymerase and the transition from the closed to the open promoter complex (Schleif 2010; Soisson et al. 1997). Additionally, in E. coli, the cAMP receptor protein (CRP) also plays an important role. According to the G. oxydans 621H genome sequences, a CRP appears to be absent (Kranz et al. 2017; Prust et al. 2005). The heterologous araC-ParaBAD system of E. coli MC4100 is very tight in G. oxydans even in the absence of AraC, indicating that promoter ParaBAD is not active in G. oxydans without AraC. The system is also well-tunable with up to 480-fold induction by l-arabinose concentrations up to 1% (w/v) (Fig. 2b). Unexpectedly, the performance of this system was strongly affected by only one or two small sequence alterations in the constructed plasmids. A single G to C exchange between the RBS and the ATG start codon of the reporter gene to create a NdeI cloning site in the MCS region of the araC-ParaBAD empty vector and/or the short remaining sequence of the XhoI site (CTCGAG) after the stop codon of the reporter gene, strongly reduced the maximum of the resulting reporter activity. In future studies, systematic analysis of such and other sequence alterations including terminators and orientations of neighboring genes (for example, target gene and the plasmid resistance cassette) may reveal specific rules that should be considered for successful construction and establishment of tight and regulatable heterologous expression systems in Gluconobacter or in AAB.
a The region of the ParaBAD promoter between the araC and araBAD genes illustrating the regulatory mode of action by the AraC regulator protein according to Schleif 2010. b AraC-dependent induction of mNeonGreen (mNG) expression up to 480-fold in G. oxydans 621H and mutant strain BP.6 both with plasmid pBBR1MCS-5-araC-PBAD-mNG induced by concentrations of l-arabinose (Fricke et al. 2020). As further analyzed and discussed in the study, the decrease of the mNG fluorescence in strain 621H at stronger induction is pH-dependent due to enzymatic oxidation of l-arabinose forming l-arabinonic acid and can be avoided by either using the multi-deletion strain BP.6 or pH-controlled condition
The l-arabinose concentrations required for highest induction in G. oxydans were much higher than the ones typically used for E. coli. Attempts to increase the arabinose sensitivity of the araC-ParaBAD system by expression of the l-arabinose transporter gene araE in tandem with araC from the promoter of araC had a strong negative effect on the growth of G. oxydans and the expression performance (Fricke et al. 2020). With a pBBR1MCS-5 derivative carrying araE separately under control of the weak constitutive G. oxydans promoter PGOX0384 no G. oxydans 621H transformants could be obtained yet, suggesting a severe growth defect upon araE expression in G. oxydans, even in the absence of l-arabinose (our unpublished results). Thus, before using AraE to hopefully increase the sensitivity of the araC-ParaBAD system toward l-arabinose, the strong negative effect of araE expression should be understood and overcome. Therefore, conditional and tuned expression of araE using the araC-ParaBAD system itself could help to clarify the negative effects of araE expression in G. oxydans. Likewise, araC-ParaBAD-dependent conditional expression could also help to express genes for other “difficult” transporters and (membrane-bound) enzymes in AAB.
Target gene expression in Komagataeibacter
Taken into account the changes in the AAB genera systematics and the associated transfers of species as described before, we found 42 studies for Komagataeibacter reporting on expression plasmids, among which 5 major lineages can be identified. Almost all studies employed electroporation as the method of choice for transfer of plasmids into Komagataeibacter, whereas conjugation was rarely used (Hall et al. 1992; Zhu et al. 2012).
Early plasmid studies in Komagataeibacter
Initial work on the genetics to provide a gene transfer system was published by Johs Kjosbakken and co-workers in 1986 when studying the conjugative transfer of broad-host-range plasmids and cloning vectors (RP1, pKT210, pRK290, pBR322) with several antibiotic resistances into bacterial cellulose-producing K. xylinus (alias Ga. xylinus alias A. xylinum) and analyzing the number of transconjugants (Valla et al. 1986; Valla et al. 1987). The plasmid RP4::Mu cts61 was used for the insertion of Tn1 into the endogenous and conjugatively mobilizable 16-, 44-, and 64-kb plasmids of K. xylinus. Many of the Tn1 insertions affected the copy number of the plasmids, yet it provided a selectable marker for the potential use of the plasmids in gene transfer experiments. In follow-up studies on cellulose-forming (Cel+) and cellulose-negative (Cel-) mutants of K. xylinus the broad-host-range cloning vector pVK100 and cosmid vector pRK311 were used to prepare gene libraries of K. xylinus to search for genes expressed from their native promoters and complementing the Cel- phenotype (Fjaervik et al. 1991; Valla et al. 1989). The screenings revealed a uridine 5’-diphosphoglucose (UDPG) pyrophosphorylase gene designated celA involved in cellulose biosynthesis for which UDPG is the precursor, and a gene encoding phosphoglucomutase activity, which catalyzes the second step of UDPG synthesis from glucose by isomerization of glucose-6-phosphate to glucose-1-phosphate.
In a similar complementation screening, a cosmid vector derived from the high copy number, double-replicon vector pKT230, exhibiting a broad host range and found to be stably maintained in K. xylinus, was used for the construction of a genomic library (Wong et al. 1990). Four genes designated bcsA, bcsB, bcsC, and bcsD, which form the cellulose synthase operon, were found to complement the Cel- phenotype caused by a cellulose synthase deficiency. In this study, it was also discovered that K. xylinus could be transformed by DNA via electroporation and a small endogenous cryptic plasmid (3.6 kb) was found. The latter was used to construct two shuttle plasmids by ligating the small cryptic plasmid with the E. coli plasmids pUC18 and pUC19 at the SstI sites to obtain pUC18-824 and pUC19-824. These shuttle plasmids confer ampicillin resistance and also serve as expression vectors due to the presence of Plac from E. coli which is actively transcribed in K. xylinus. Cloning of the single bcs genes expressed from Plac allowed specific complementation of different Cel- mutants. Additionally, replacing the native bcs promoter Pbcs on the chromosome with the heterologous promoters Plac or Ptac reduced the cellulose synthase activities of cells to -60% and -85% of the level of the parental strain, demonstrating that the promoter Pbcs is quite strong and useful for high expression of desired target genes. In a later study on the cellulose-synthesizing operon acs in another K. xylinus strain, the cosmid vector pRK311 already mentioned above was used to express the single gene acsD from Ptac for recovery of normal cellulose production in an acsD mutant (Saxena et al. 1994).
pUF106, pTA99, and pTI99—a first major expression plasmid lineage
The principle of fusing endogenous cryptic plasmids derived from K. xylinus strains with well-characterized E. coli plasmids to obtain shuttle vectors was also tested and applied in further studies using pUC18, pBR322 or the pBR322-based plasmid pTrc99A. Based on the endogenous K. xylinus plasmid pFF6 (2.72 kb) fused with pUC18 or with pTrc99A, the plasmids pUF106 and pTA99 were constructed to obtain shuttle vectors (Fujiwara et al. 1992; Tajima et al. 2001). Plasmid pTA99 was used to express the (exo)polysaccharide metabolism-related β-glucosidase gene bglxA from a K. xylinus strain driven by the strong E. coli Plac-Ptrp hybrid promoter Ptrc. Analysis of culture supernatants demonstrated an approximately tenfold higher specific β-glucosidase activity compared to K. xylinus cells carrying the empty vector as a control. Similar to pTA99, shuttle vector pTI99 was also obtained by fusion of pFF6 with pTrc99A (Hu et al. 2010). It was used to clone various gene variants encoding N-terminally truncated AxCeSD proteins that form subunit D of the cellulose-synthesizing terminal complex in K. xylinus (Hu et al. 2010). Thereby, the N-terminal loop of subunit D, especially residue Lys6, turned out to be important for cellulose production. The AxCeSD and CcpAx proteins of the cellulose-synthesizing terminal complex were also expressed as fusion proteins with an enhanced green fluorescent protein (EGFP) using pTI99 (Sunagawa et al. 2013). In fluorescence microscopy analysis, the AxCeSD-EGFP fusion protein showed a cellular localization similar to the CcpAx fusion protein. Together with other data, AxCeSD and CcpAx showed significant interaction and were suggested to function as members of the terminal complex in K. xylinus.
To obtain modified bacterial cellulose with altered mechanical strength, biodegradability, and bioactivity for biomedical use, the curdlan synthase gene crdS from Agrobacterium was expressed in K. xylinus using pTI99 (Fang et al. 2015). It enabled curdlan (β-1,3-glucan) to be synthesized in K. xylinus simultaneously with cellulose nanofibers in vivo for biopreparation of nanocomposites. Production of bacterial cellulose with altered and advantageous properties could also be obtained without chemical modifications solely through altering the tight spatial organization of the cellulose fibers using a non-motile cellulose-producing K. hansenii with increased cell length and the ability to move on the surface of the medium (Jacek et al. 2019; Jacek et al. 2018). Therefore, the motAB genes encoding motor and stator proteins essential for flagellum rotation in numerous bacterial species were expressed either as an operon, or alone, or each gene as a translational fusion with gfp using the pTI99 vector backbone. It was assumed that probably the torque produced by the MotAB proton pump could affect other yet unknown motility mechanisms, cell division, filamentation or transport, thereby affecting the structure of the cellulose. Indeed, K. hansenii mutants with increased cell length and motility were shown to produce altered cellulose membranes with increased pore sizes and a relaxed fiber structure, which supported chondritic cell proliferation important for potential future application in tissue engineering.
pSA19—a second major expression plasmid development
In parallel to the pFF6-based plasmid lineage, another endogenous cryptic plasmid termed pAH4 (4 kb) from a cellulose-producing K. xylinus strain was used for construction of a shuttle vector and established pSA19 as another major expression plasmid. HindIII-linearized pAH4 and pUC18 were fused to obtain the shuttle vector pSA19, which contains several pUC18 cloning sites and the promoter Plac for expression of cloned genes (Tonouchi et al. 1994). The copy number of pSA19 in K. xylinus is roughly ten per genome and transformation efficiency was strongly increased when recombinant pSA19 plasmids were propagated in K. xylinus instead of E. coli, suggesting the presence of an effective restriction-modification system in K. xylinus. Later, restriction data obtained from two cryptic plasmids discovered in K. xylinus B42 showed that these plasmids contain protected EcoRI and ApoI sites. The protection was also present in the chromosomal DNA and the results suggest the presence of a modification system in K. xylinus that recognizes the tetranucleotide 5’-AATT (Petroni et al. 1996). However, plasmid pSA19 was successfully used in 8 further studies on cellulose-producing K. xylinus strains:
-
(i)
pSA19 was used to express a wild-type endo-β-1,4-glucanase from Bacillus and a mutated variant (H131F), which is inactive but still binds to cellulose, to study their effects on cellulose production by K. xylinus (Tonouchi et al. 1995). The native glucanase enzyme enhanced cellulose production and reduced the amount of a polysaccharide called acetan, while the inactive variant did not affect cellulose production. It was concluded, therefore, that the endoglucanase activity itself, but not the cellulose-binding ability, was essential for the enhancement of cellulose production.
-
(ii)
The extracellular sucrase gene sucZE3 from Zymomonas mobilis together with the secretion-activating factor gene zliS and the B. subtilis levansucrase gene sacB containing a mutation causing decreased levan-forming activity were cloned into pSA19 under the control of Plac to study cellulose production by Komagataeibacter from sucrose, which is the most suitable carbon source for the economical production of bacterial cellulose (Tonouchi et al. 1998b). The gene expression resulted in increased cellulose production and reduced levan accumulation.
-
(iii)
A sucrose phosphorylase (SPase) gene from Leuconostoc mesenteroides was expressed with pSA19 using several promoters (Plac, Ptac, Pugp from K. xylinus, Pgdh from G. oxydans) to improve cellulose production in K. xylinus (Tonouchi et al. 1998a). Compared to expression from Plac, the SPase expression level was 78% with Ptac, 37% with Pugp, and only 13% with Pgdh. Interestingly, a small increase of the 5’-UTR length with a modified Plac region increased the SPase expression level 3-fold.
-
(iv)
pSA19 was used to express a sucrose synthase gene with the 5’-upstream region (~3.1 kb) of the cellulose synthase operon bcs from a K. xylinus strain instead of Plac to study this bcs promoter region (Nakai et al. 1998). In K. xylinus, the expression occurred more than 241 bp upstream from the ATG start codon within the 1.1 kb upstream region. In A. aceti the expression was 75% of that in K. xylinus, while in E. coli no expression at all was detected, suggesting that the bcs upstream region studied may function as an ABB-specific promoter.
-
(v)
To make use of the free energy of the glycosidic bond in sucrose for cellulose biosynthesis, the gene encoding native sucrose synthase from mung bean or a variant with a S11E mutation mimicking phosphorylation were cloned and expressed with pSA19 in K. xylinus under control of Plac (Nakai et al. 1999). Sucrose synthase reversibly converts sucrose and UDP to fructose and UDP-glucose, the substrate of cellulose synthase. Expression of sucrose synthase in K. xylinus enhanced cellulose production from sucrose and the S11E variant with an increased sucrose affinity had an even stronger stimulating effect on cellulose synthesis.
-
(vi)
For complementation of an ORF2 disruptant mutant of K. xylinus and functional analysis of the ORF2 polypeptide involved in cellulose synthesis, the ORF2 sequence plus a kanamycin resistance cassette was cloned into pSA19 (Nakai et al. 2002). The parental strain produced tough, colorless, and insoluble cellulose pellicles, whereas the ORF2 mutant produced thin, yellow, and fragile pellicles. The ORF2 polypeptide was suggested to be involved in the assembly of glucan chains into crystalline cellulose I microfibrils.
-
(vii)
For complementation of K. xylinus mutants with disrupted aceR and aceQ, these genes involved in acetan biosynthesis were cloned into pSA19 for expression (Ishida et al. 2002). NMR and ESI-MS analyses of the produced water-soluble polysaccharides suggested that aceQ and aceR encode a glucosyltransferase and a rhamnosyltransferase, respectively.
-
(viii)
In a mutant of K. xylinus in which the carboxymethylcellulase gene cmcax was disrupted by an ampicillin resistance cassette, the cmcax gene was expressed in trans using the pSA19 backbone carrying a kanamycin resistance cassette as an additional selection marker (Nakai et al. 2013).
pBBR122 and pMV24—two other major expression plasmids used in Komagataeibacter
Another expression plasmid stably used in Komagataeibacter is the broad-host-range plasmid pBBR122 introduced above in the Gluconobacter section. With this plasmid, the gene of the bacterial hemoglobin from Vitreoscilla (VHb) was expressed driven by the constitutive bla promoter in cellulose-producing K. xylinus (Chien et al. 2006; Liu et al. 2018; Setyawati et al. 2007). VHb has been widely applied to improve cell survival during hypoxia. The hemoglobin was biochemically active also in K. xylinus and enhanced both the growth rate in shaken cultures by 50% and the cellulose titers. VHb allowed growth or survival of K. xylinus at lower oxygen tension and facilitated cellulose production in static culture, in which the polysaccharide exhibited interesting altered material properties. Cellulose nanofibers can also be used to self-immobilize K. xylinus in a biofilm, with the advantage of better resistance toward harsh biotransformation reaction conditions. This was tested for the production of α-ketoacids by a heterologous d-amino acid oxidase (DAAO), the gene of which was constitutively expressed from Plac using pBBR122 (Setyawati et al. 2009). In the DAAO-catalyzed reaction toxic H2O2 is formed as a product. With self-immobilized K. xylinus cells expressing the DAAO gene, the system exhibited improved thermal and operational stability, as well as easy retrieval for repeated use. In contrast, for biomedical and biomass conversion applications, degradability of bacterial cellulose is important. To improve the poor in vitro and in vivo degradability of bacterial cellulose, a three-gene operon from Candida albicans for UDP-N-acetylglucosamine (UDP-GlcNAc) synthesis was expressed from the constitutive promoter Pbla in K. xylinus with the aim to introduce GlcNAc residues by cellulose synthase and produce a chimeric polymer (Yadav et al. 2010). The modified bacterial cellulose exhibited a high GlcNAc content and lower crystallinity, making it a multifunctional bioengineered polymer susceptible to lysozyme, an enzyme widespread in the human body, and to rapid hydrolysis by cellulase, an enzyme commonly used in biomass conversion.
Another shuttle vector used in Komagataeibacter studies from 2008 onward is pMV24 already constructed and introduced in 1989 for expression in Acetobacter (see Acetobacter section below). pMV24 was used to study the regulation and function of the genes ginI, ginA and ginR of a quorum-sensing system from a K. intermedius strain producing three different N-acylhomoserine lactones (Iida et al. 2008a). The data demonstrated that the GinI/GinR quorum-sensing system controls the expression of ginA, which in turn inhibits oxidative fermentation, including acetic acid and gluconic acid fermentation by an unknown mechanism. In a further study, it was discovered that expression of the outer membrane protein gene gmpA is positively controlled by the GinA protein in an unknown manner (Iida et al. 2008b). Complementation studies with the expression of gmpA using pMV24 demonstrated that GmpA plays a role in inhibiting the formation of oxidized products, including acetic acid and gluconic acid. Transcriptome analysis revealed gltA encoding a putative glycosyltransferase, pdeA encoding a putative cyclic-di-GMP phosphodiesterase, and nagA encoding a putative N-acetylglucosamine-6-phosphate deacetylase as further target genes whose expression is influenced by the GinA protein. For functional and phenotypic analysis gltA and nagA were expressed under the control of both their own promoter as well as the promoter Plac of pMV24 and pdeA was expressed from Plac in K. intermedius (Iida et al. 2009). Recently, pMV24 was used to express the acyl-CoA dehydrogenase (ACAD) gene caiA from a thermotolerant K. intermedius in a non-thermotolerant K. medellinensis strain (Konjanda et al. 2019). It improved growth, acetic acid tolerance and ethanol oxidation even at higher temperature.
Other pBR322-based expression plasmids used in Komagataeibacter
Another pBR322-based plasmid is pUCD2 originally developed for Agrobacterium and exhibiting autonomous replication and an active Ptet promoter of the tetC gene for tetracycline resistance in K. hansenii (Deng et al. 2013). pUCD2 was used to test the complementation of K. hansenii Cel- mutants by expressing the cellulose synthase complex gene acsA, the guanylate dicyclase gene dgc1, and the putative transcriptional regulator gene crp-fnr (ccp) from their native promoters (Deng et al. 2013). In this study, pUCD2 with a promoterless tetC gene was also used to test promoter activities in E. coli and K. hansenii to study the acs operon and ccp promoter regions. pUCD2 was also used for complementation by expressing lysine decarboxylase and alanine racemase genes fused at the 3’ ends (C-terminally) with the sequence for an octapeptide FLAG epitope tag allowing detection via immunoblotting (Deng et al. 2015). Furthermore, pBR322 was used to create the shuttle vectors pBE2 and pBE3 by ligating linearized native pGE2 and pGE3 plasmids from K. europaeus with pBR322 (Akasaka et al. 2015). pBE2 and pBE3 can replicate in K. europaeus, but were not used further. Another pBR322-based vector is pCTP1 containing a chlamydial plasmid cloned into pBR322 and used to express acsD of the cellulose synthesizing operon acs in fusion with gfp from Plac to produce N-terminal and C-terminal fusion proteins in complementation studies of a K. xylinus acsD disruption mutant (Mehta et al. 2015). The data obtained suggested that the AcsD protein aids in the proper orientation of the linear terminal complexes along the longitudinal axis of the cell, thus AcsD is involved in the final stage of the hierarchical assembly of cellulose.
pSEVA plasmids and regulatable expression in Komagataeibacter
The most recent lineage of expression plasmids used in Komagateibacter is based on the Standard European Vector Architecture (SEVA) toolkit, a resource for constructing optimal plasmid vectors based on a minimized backbone and three interchangeable modules to design a compilation from several origins of replication, diverse antibiotic resistance markers, and a cargo of interest, flanked by uncommon restriction sites (Durante-Rodriguez et al. 2014). Based on SEVA, a complete set of tools was developed for engineering Komagataeibacter that consists of protocols, modular plasmids, promoters, reporter proteins, and inducible constructs that should enable external control of gene expression (Florea et al. 2016a). Eight SEVA-based plasmids with different ori and low, medium or high expected copy number were assessed for propagation in K. rhaeticus iGEM. With ori RK2, pBBR1, and RSF1010 also mentioned above or below and pWV01, the respective SEVA plasmids were found to show replication, while with ori R6K, pRO1600/ColE1, pMB1 and ColE1/pBR322 the SEVA constructs tested did not show replication. The latter, ori pBR322 could be used in Komagataeibacter at least in other plasmids as mentioned above. From seven reporter proteins tested, mRFP1, GFPmut3, and sfGFP showed visually detectable expression. With mRFP1 as reporter 10 promoters from an open-access collection of synthetic minimal E. coli promoters were tested for their promoter strength in K. rhaeticus iGEM. For inducible gene expression, four constructs expected to be induced externally by anhydrotetracycline (ATc) or N-acyl homoserine lactone (AHL) were tested. From these, the AHL-inducible constructs showed higher induction and lower leakiness than the ATc-induced constructs, yet the induction fold-changes were only up to 5-fold under the conditions tested (Florea et al. 2016a). Interestingly, the LuxR-dependent AHL-inducible system showed a much better induction performance due to a very low basal mRFP1 signal when non-induced and a very strong mRFP1 signal when cells were induced inside cellulose pellicles. Furthermore, the E. coli Hfq protein that binds small regulatory RNAs (sRNAs) and mRNAs to facilitate mRNA translational regulation and a sRNA targeting UDP-glucose pyrophosphorylase (UGPase) mRNA were co-expressed from a SEVA plasmid with pBBR1 ori in response to the AHL inducer to inhibit UGPase mRNA translation. UGPase catalyzes the production of UDP-glucose, the substrate for cellulose synthesis, and is encoded by a single gene in the genome, allowing knockdown by a single sRNA. This system was found to be highly efficient, as cellulose production was suppressed completely upon full induction and could be fine-tuned using different concentrations of AHL. With E. coli Hfq and broad-host-range backbone, the system was engineered to be a general platform for targeted knockdowns in Komagataeibacter and other bacterial species independent from the host Hfq by adding new sRNA sequences to the plasmid making the construct easily modifiable for other mRNA targets (Florea et al. 2016a).
The pWV01 ori found to be functional in the above study with a SEVA backbone is also present in the non-SEVA plasmid backbone pBAV1C containing the l-arabinose-inducible araC-ParaBAD system, which was used to induce expression of the bcs operon genes bcsA, bcsAB, and bcsABCD in K. xylinus for engineering and characterization of bacterial nanocellulose films as low cost and flexible sensor material (Mangayil et al. 2017). Despite several attempts, the pBAV1C derivative with pWV01 ori could not be isolated from the transformed K. xylinus cells suggesting instability, although clear phenotypic differences were detected in growth curves and cellulose production in the presence of the plasmid.
The SEVA-based luxR-Plux system mentioned above was extended by including the AHL-synthesis gene luxI downstream of the synthetic constitutive promoter J23104 on a separate plasmid to establish synthetic cell-to-cell communication in K. rhaeticus (Walker et al. 2019). Expression of luxI allowed the production of the diffusible AHL molecule N-(3-oxohexanoyl) homoserine lactone in the transformed K. rhaeticus strain (sender). In another K. rhaeticus strain carrying the luxR-Plux system (receiver), the signal molecule was sensed, affecting the expression of the mRFP gene encoding the fluorescence reporter protein. It was demonstrated that communication can occur both within and between growing pellicles in mixed cultures of the two strains.
SEVA-based plasmids were also used to characterize 11 constitutive promoters, 3 inducible promoters (Plux, Ptet, ParaBAD), natural and synthetic terminators and ribosome binding sites as well as protein degradation tags in K. xylinus, K. hansenii, and K. rhaeticus iGEM (Teh et al. 2019). Plux was found to be stronger and less leaky than Ptet and ParaBAD. Due to the high leakiness, induction fold changes of Ptet and ParaBAD were rather small and in the case of ParaBAD depended on the carbon source supplemented for growth. In this study, CRISPR interference (CRISPRi) with a catalytically inactive Streptococcus pyogenes Cas9 protein (dCas9) was also tested as a tool to readily knock down the expression of a target gene by blocking transcription. Therefore, the dCas9 gene fused to a 3xFLAG tag DNA sequence was expressed by the strong promoter J23104 together with a single guide RNA (sgRNA) gene under the control of the native tracrRNA promoter from S. pyogenes. Alternatively, the sgRNA gene was positioned without promoter just downstream of the dCas9-3xFLAG gene, thus forming an operon. The mentioned constructs were cloned into a single pSEVA331Bb plasmid backbone. When targeting the 5’-ends of the acsAB gene and of the acsD gene of the endogenous acs operon for cellulose synthesis in K. hansenii by two different sgRNAs, a more than 2-fold decrease in acsAB expression and 15% reduced cellulose yield were observed, while for acsD no significant change in expression was observed.
pSEVA-based CRISPRi was also used in K. xylinus to control the expression level of galU encoding UGPase controlling the metabolic carbon flux between the cellulose synthesis pathway and the pentose phosphate pathway (Huang et al. 2020). By overexpression of galU and interfering with different sites of the galU sequence using CRISPRi, varying expression levels of galU from 3 to 3000% were obtained. Analysis of the cellulose characteristics showed that porosity was negatively and crystallinity was positively correlated with increasing galU expression levels. This results also confirmed that the properties of the bacterial cellulose can be altered without chemical modifications to increase the application potential in different fields. Furthermore, these studies showed that CRISPRi as well as Hfq-mediated RNA interference can be used to modulate target gene expression in Komagataeibacter and potentially also in other AAB.
To enhance bacterial cellulose production by K. xylinus, the effect of the expression levels of the UGPase gene galU, the phosphoglucomutase gene pgm, and the nucleoside-diphosphate kinase gene ndp were analyzed using pSEVA331 derivatives and synthetic RBSs exhibiting different strength identified via fluorescence-activated cell sorting (FACS) in a GFP reporter library (Hur et al. 2020). With pSEVA331-based expression of all mentioned genes under the control of PJ23104 and with a selected synthetic RBS, the bacterial cellulose titer was 4-fold higher under shaking conditions (3.7 g/L) than that of wild-type K. xylinus. In static conditions 5.3 g/L cellulose was reached, demonstrating that reinforced metabolic flux toward bacterial cellulose through modified gene expression represents a practical approach for the improvement of bacterial cellulose production. By another strategy, which included the deletion of a PQQ-dependent glucose dehydrogenase gene and co-expression of the glucose facilitator gene glf from Zymomonas mobilis and the endogenous glucokinase gene glk from K. xylinus, a somewhat higher cellulose titer (5.9 g/L) was obtained with K. xylinus in a specific growth condition (Liu et al. 2020b). In this study, expression plasmid pRedGX with a pBBR1 ori, a kanamycin resistance gene kanR, lacIq, and the λ-red gene controlled by LacIq-dependent Ptrc was constructed via Gibson assembly in a SEVA-like manner. This plasmid was used for the IPTG-induced expression of λ-Red recombinase gene in order to increase the efficiency of homologous recombination between the targeted chromosomal gene-flanking regions and a linear PCR product consisting of the in-frame deleted gene and chloromycetin resistance gene with flanking FLP recognition target (FRT) sites, all flanked by the targeted gene-flanking regions. A second expression plasmid, pFLPGX, containing lacIq, a pBBR1 ori flanked by FRT sites, a spectinomycin resistance gene, and the FLP recombinase gene flp controlled by LacIq-dependent Ptrc, was also constructed via Gibson assembly. pFLPGX with the same ori as pRedGX but a different resistance gene was used to eliminate the λ-red plasmid by antibiotics selection and to produce FLP recombinase, which catalyzes the reciprocal recombination between the FRT sites introduced into the chromosome for in-frame gene deletion and also between the FRT sites in the flp plasmid (FRT-pBBR1 ori-FRT) for self-elimination of the flp plasmid. Using the λ-red and the flp expression plasmids, four putative glucose dehydrogenase genes were deleted in K. xylinus, resulting in the identification of one of these genes as being responsible for gluconate formation from glucose. In the respective mutant, the glf and glk genes were constitutively co-expressed from a plasmid with a pBBR1 ori and a kanamycin resistance gene. The results obtained indicated that glucose was transported into the cell by the facilitator protein and glucokinase further increased the production of cellulose (Liu et al. 2020b). In contrast, the constitutive expression of the glucose phosphotransferase system genes ptsG and ptsHIcrr from E. coli did not significantly increase the efficiency of glucose utilization, likely because of limited availability of PEP, which is the donor of the phosphate group during PTS-catalyzed glucose uptake.
Genomic integrations for target gene expression in Komagataeibacter
In two studies, genomic integrations instead of plasmids have been used for the expression of target genes in Komagataeibacter. A wild-type K. xylinus strain was modified by random transposon mutagenesis to insert the E. coli β-galactosidase gene lacZ generating a lactose-utilizing and cellulose-producing mutant strain (Battad-Bernardo et al. 2004). The promoterless lacZ gene expressed from the Tn10 cassette inserted once into the chromosome was constitutively expressed, alleviated the growth retardation in lactose medium and was stably maintained in a non-selective medium for more than 60 generations. The modified strain showed a 28-fold increase in cellulose production when grown in lactose medium and could utilize 17 g/L of lactose in whey substrate within 4 days. In a genomic and metabolic analysis of a K. xylinus strain producing bacterial cellulose nanofiber (CNF), glucose-6-phosphate isomerase and 6-phosphogluconate dehydrogenase encoded by pgi and gnd have been predicted as novel overexpression targets for the enhanced CNF production (Jang et al. 2019). Therefore, the heterologous pgi and gnd genes from E. coli and Corynebacterium glutamicum were individually constitutively overexpressed from the chromosomal sacB locus under the control of Ptac. By this approach, the amount of CNF produced in a complex medium containing glucose was doubled compared to that obtained with the control strain.
Target gene expression in Acetobacter
For Acetobacter, we found 30 publications reporting on recombinant DNA work with expression plasmids and 2 major lineages can be seen. The first studies were published in 1985 by Teruhiko Beppu and co-workers. Here, the first chimeric shuttle vectors for E. coli / A. aceti (Acetobacter subgenus Acetobacter) have been constructed by ligation of cryptic plasmid pMV102 endogenously present in A. aceti subsp. xylinum NBI 1002 with the E. coli plasmids pACYC177, pBR322, or pBR325. The initial size of chimeric plasmids (>6 kb) was reduced by a series of consecutive digestion and ligation steps delivering pMV329 with only 3.4 kb as the smallest vector (Fukaya et al. 1985a). Most of the constructed shuttle vectors were stably maintained in Acetobacter. In parallel, improvements in the chemically induced competence for the transformation of Acetobacter were reported (Fukaya et al. 1985c; Okumura et al. 1985). Furthermore, shuttle vectors pMVL1 and pMVL2 were constructed by ligation of the pMV102 plasmid with a pBR322 derivative containing the β-isopropylmalate dehydrogenase gene leuB from A. aceti under control of its native promoter (Okumura et al. 1988). Therefore, pMVL1 and pMVL2 carrying an ampicillin resistance cassette were the first A. aceti / E. coli shuttle vectors with double selection markers as the leuA gene allowed selection of leu+ transformants in an A. aceti leuA- host. Both pMVL1 and pMVL2 appeared to be stably maintained in A. aceti as plasmids after 4 times of successive inoculation and cultivation.
pMV24—the first major expression plasmid used in Acetobacter
The smallest plasmid of the pMV series described above (pMV329; 3.4 kb) was used to construct the shuttle vector pMV24 by inserting pMV329 between the AatII and NdeI sites of pUC18 (Fukaya et al. 1989). Plasmid pMV24 exhibits an estimated copy number of 10 in A. pasteurianus, promoter Plac, and allows translational fusion of the target protein with the 10 N-terminal amino acids of the E. coli β-galactosidase. pMV24 was used for recombinant DNA work in 13 Acetobacter studies for expression of various genes from the Plac promoter present on the plasmid and additionally from the native gene promoter, if present on the cloned insert:
-
(i)
A membrane-bound aldehyde dehydrogenase ALDH from A. polyoxogenes fused to the short N-terminal β-galactosidase peptide was produced in an A. aceti mutant lacking the enzyme activity (Fukaya et al. 1989). In submerged fermentation, expression of the ALDH-encoding gene caused an approximately 2-fold increase of the production rate and the maximum concentration of acetic acid.
-
(ii)
Complementation studies with acetic acid-sensitive mutants of A. aceti obtained by chemical mutagenesis revealed three genes termed aarA, aarB, and aarC responsible for acetic acid resistance. Gene aarA was identified to encode a citrate synthase (Fukaya et al. 1990).
-
(iii)
The aarC gene expressed from the pMV24 backbone was found to functionally complement an aarC gene disruptant mutant of A. aceti, which is defective in acetate assimilation (Fukaya et al. 1993).
-
(iv)
A part of the promoter region of the membrane-bound alcohol dehydrogenase (ADH) gene of A. pasteurianus was cloned into pMV24 to corroborate a second transcriptional start by primer extension analysis (Takemura et al. 1993b). Furthermore, a promoterless chloramphenicol acetyltransferase (CAT) gene was cloned into pMV24 resulting in pMVC18 usable for promoter analysis by CAT activity assays. It was shown that the more than 10-fold increased ADH activity caused by ethanol was not due to increased transcription of the adh gene, suggesting a mechanism involving translational or posttranslational regulation.
-
(v)
In a search for genes conferring ethanol resistance, an ethanol-sensitive A. pasteurianus mutant was transformed with a pMV24-based genomic library and the recombinants were screened for ethanol resistance. The his1 gene encoding histidinol phosphate aminotransferase was shown to be responsible for the suppression of the ethanol sensitivity of the mutant. This effect was dependent on the higher copy number of his1 obtained with plasmid pMV24, as the single genomic his1 copy was not sufficient to enable ethanol resistance (Takemura et al. 1993a).
-
(vi)
The adhS gene encoding subunit III of the three-component membrane-bound alcohol dehydrogenase (ADH) from A. pasteurianus was expressed from pMV24 and restored the ADH activity of an adhS mutant deficient in subunit III (Kondo et al. 1995).
-
(vii)
In a follow-up study, the gdhS gene for subunit III of the ADH from G. suboxydans, which strongly differs in its amino acid sequence from GdhS of A. pasteurianus, was expressed in the A. pasteurianus adhS mutant and could not restore the ADH activity (Kondo and Horinouchi 1997). (viii) A study on the esterase-encoding genes est1, which is induced only in the presence of ethanol, and est2, which is repressed in the presence of ethanol, suggested a cis-acting domain exerting transcriptional regulation in the 5’-region of est2 mRNA (Kashima et al. 1999). Furthermore, the est1 promoter activity was analyzed using the CAT reporter plasmid pMVC18 introduced above to demonstrate the in vivo effect of ethanol on the transcription of est1.
-
(viii)
In a follow-up study, est1 was shown to play a major role in ethyl acetate formation and its production was fully recovered in an A. pasteurianus est1 mutant by expressing an est1-containing DNA fragment from pMV24 (Kashima et al. 2000).
-
(ix)
In A. aceti, pMV24-based overexpression of groESL encoding the chaperones GroEL and GroES, which are induced not only by temperature shifts but also by other stresses such as exposure to ethanol, conferred improved growth of A. aceti at increased temperature (42°C) and also supported the resistance against ethanol stress (Okamoto-Kainuma et al. 2002).
-
(x)
Similarly, pMV24-based overexpression of the molecular chaperone operon dnaKJ with its heat-shock promoter resulted in improved growth of A. aceti compared to the control strain at high temperature or in the presence of ethanol, suggesting a correlation to resistance against various stressors present during fermentation, although it did not increase the resistance to acetic acid (Okamoto-Kainuma et al. 2004).
-
(xi)
Acetic acid resistance of A. aceti was increased when the aconitase gene was overexpressed from Plac using pMV24, showing that multiple copies of the aconitase gene, and accordingly an increase in aconitase activity, conferred acetic acid resistance (Nakano et al. 2004).
-
(xii)
In A. pasteurianus, pMV24-based overexpression of the uvrA gene, whose product is involved in recognition and processing of DNA lesions, resulted in increased biomass formation in the presence of high acetic acid concentrations and resulted in upregulation of enzymes involved in ethanol oxidation and acetic acid tolerance. UvrA thus contributes to acetic acid tolerance by protecting genome integrity, thereby ensuring gene expression and metabolism (Zheng et al. 2018).
pCM62—the second major expression plasmid used in Acetobacter
In other studies with Acetobacter, the improved broad-host-range cloning vector pCM62 was used as an expression plasmid. It was originally developed for use in methylotrophs and other Gram-negative bacteria by combining the functions present in the minimal transferable and selectable RK2 replicon of the pTJS75 derivative pCM51 with the polylinker and pBR322 ori of pUC19 (Marx and Lidstrom 2001). pCM62 was used to express from the promoter Plac or the native promoter, if present on the inserted fragment, a number of different genes:
-
(i)
The UDP-galactose synthesis gene galE was expressed for complementation of a galE mutant of A. tropicalis, which was found to produce extracellular polysaccharide secreted into the medium instead of forming capsular polysaccharide involved in pellicle formation (Deeraksa et al. 2006).
-
(ii)
The subunit III gene adhS of the PQQ-dependent alcohol dehydrogenase (ADH) from A. pasteurianus was expressed to complement an adhS disruptant mutant, which revealed that subunit III is essential for the formation of the active ADH complex besides subunit I and II (Masud et al. 2010).
-
(iii)
Several selected genes identified to support growth of A. tropicalis at higher temperatures were expressed to complement gene disruptant mutants obtained by transposon mutagenesis (Soemphol et al. 2011). The study revealed a serine protease, glutamine synthetase, lysyl-tRNA synthetase, 3-phosphoglycerate dehydrogenase, and a few other proteins to rescue the growth defect of the respective mutant strain at higher temperature (42°C).
-
(iv)
The chaperone genes groESL were expressed in an A. pasteurianus groEL disruptant mutant for complementation and in the parental strain, which enhanced the resistance against 4% acetic acid and 5% ethanol (Theeragool et al. 2018).
-
(v)
The membrane-bound aldehyde dehydrogenase genes aldFGH, which are indispensable for acetic acid formation from ethanol, and the isozyme genes aldSLC, whose mRNA levels were 10-fold lower during growth on ethanol compared to growth without ethanol, were expressed in A. pasteurianus (Yakushi et al. 2018a).
-
(vi)
A mutated oxidoreductase gene fabG that may change the fatty acid composition and supports growth of A. pasteurianus in a low-nutrient environment were expressed. It enhanced growth of A. pasteurianus and the acetic acid production useful for cost-effective manufacturing of rice vinegar (Phathanathavorn et al. 2019).
Other expression plasmids used in Acetobacter
Other shuttle vectors for cloning were prepared using the replication region of the large native ~19 kb plasmid pAC1 from A. pasteurianus and the kanamycin or tetracycline resistance markers obtained from pUC4-KAPA and pBR322 fragments to yield pACK1 (4.5 kb), pACT7 (3.9 kb) and its smaller BalI / PvuII-digested and self-ligated derivative pACT71 (3.3 kb) (Grones and Turna 1992). Plasmid pACT7 was further modified to include a kanamycin resistance cassette enabling double selection, resulting in plasmid pACG3 (4.8 kb). These shuttle vectors for E. coli / A. pasteurianus exhibit only the replication region from pAC1, yet possess a similar ability to replicate in E. coli and A. pasteurianus as the shuttle vectors of the pMV series developed for A. aceti mentioned above. The plasmids with pAC1 replicon only were successfully expressed in twelve Acetobacter species and transfer of plasmid DNA was optimized for A. pasteurianus (Bilska and Grones 2003; Grones and Turna 1995). The pAC1 replicon only plasmid pACT71 was used to express the E. coli β-galactosidase gene from Plac in A. pasteurianus to analyze its production and release of the protein through the outer membrane in dependence of the medium composition (Grones and Bencova 1994).
In a study on the functions of the multiple heme c moieties in intramolecular electron transport and ubiquinone reduction by ADH, the expression plasmid pKS13 providing tetracycline resistance was used to express the alcohol dehydrogenase (ADH) genes adhAB from A. aceti in A. pasteurianus NP 2503 deficient in ADH (Matsushita et al. 1996). Later, the two cloning and expression vectors pMK10 and pMK20 were constructed from the small A. pasteurianus plasmid pAP1 (3 kb) conferring kanamycin resistance, which can also replicate in E. coli (Kretova and Grones 2006). Plasmid pMK10 was constructed from a pAP1 fragment containing the origin and the HaeII fragment of pUC19 carrying the lacZ’ gene. Plasmid pMK20 was constructed from another pAP1 fragment with the origin fused with the EcoRI-PstI fragment of plasmid pCE30 carrying the PL and PR promoters and the gene encoding the temperature-sensitive repressor cI857 from bacteriophage lambda. The plasmids pMK10 and pMK20 were stable 96–98% both in A. pasteurianus and E. coli after 250 generations in the absence of kanamycin. Plasmid pMK20 was successfully used for cloning and expression of a membrane protein from Salmonella (unpublished), but no further work based on these plasmids was reported.
In one Acetobacter study, the IPTG-inducible PT7-based Novagen vector pET-30a(+) with pBR322 origin was described to have been used in A. pasteurianus for inducible expression of the endogenous genes of DnaA- and DnaB-like proteins as well as the dnaA and dnaB genes from E. coli to study the replacement of the corresponding endogenous proteins expressed from the chromosome (Bugala et al. 2016). A strong induction of the plasmid-based gene expression in A. orleanensis was reported, yet it was not explained whether it resulted from the T7 promoter present on pET-30a(+) or from another promoter included by the cloning procedure. Furthermore, plasmid backbone pBAD18 carrying pBR322 origin, the classical l-arabinose-dependent AraC regulator gene araC and the ParaBAD target promoter of AraC have been used in A. orleanensis for arabinose-inducible expression of asRNA fragments (121 nucleotides) which are antisense to the C-terminal coding sequence of the dnaA and dnaB gene sequences studied (Bugala et al. 2016). The data of the asRNA induction by arabinose have not been shown, thus a conclusion about leakiness and induction performance is not possible yet.
In two recent Acetobacter studies, the pBBR1-based broad-host-range cloning vectors pBBR1MCS-4 and pBBR1MCS-2 already introduced above in the Gluconobacter section were used. pBBR1MCS-4 was used to express in A. pasteurianus the adhAB operon of the endogenous PQQ-dependent alcohol dehydrogenase (ADH) from its native promoter Padh (Wu et al. 2017). By this, the acetic acid production was improved while the residual ethanol content was decreased. 2D proteomic analysis indicated that 19 proteins with several functional classifications were differently expressed more than 2-fold upon adhAB overexpression. Metabolic flux analysis of the pathway from ethanol and glucose revealed that overproduction of the PQQ-dependent ADH is an effective way to improve the ethanol-oxidizing respiratory chain. pBBR1MCS-2 was used in A. pasteurianus to express genes of the membrane-bound alcohol dehydrogenase (adhA) and aldehyde dehydrogenase (aldH), PQQ biosynthesis genes (pqqAB or pqqABCDE), and combinations thereof, each under the control of the promoter Ptuf that was 1.8-fold stronger than PadhA in GFP reporter assays (Gao et al. 2020a). Synergistic expression of these genes could not only efficiently relieve the conflict between increased acetic acid production (69 g/L in semi-continuous cultivation) and compromised cell fitness, but also enhanced the acetic acid tolerance of A. pasteurianus to a high initial concentration of 3% (v/v) thereby shortening the duration of the starting-up process from 116 to 99 h. This strategy is of significance for decreasing costs for producing high-strength acetic acid industrially and will also be useful for the production of other desired organic acids, especially those involving PQQ-dependent enzymes.
Target gene expression in Gluconacetobacter
Plasmid diversity in Gluconacetobacter
Taking into account the AAB genera and strain updates, as well as our exclusion criteria mentioned before, we found 10 studies using diverse plasmids for gene expression in Ga. diazotrophicus. Mostly, electroporation was used to transfer the recombinant DNA, yet conjugation was also used in some cases. The first recombinant DNA study in Gluconacetobacter was a complementation experiment in a levan synthesis-deficient mutant of the renamed A. diazotrophicus (Arrieta et al. 1996). Here, the plasmid pPW12, a derivative of the broad-host-range cosmid cloning vector pLAFR1 conferring tetracycline resistance and derived from pRK290 with RK2 ori, was used to construct a cosmid library from total genomic DNA of the parental strain to express endogenous complementing genes from their native promoters. This approach revealed a gene termed lsdA encoding a levansucrase potentially interesting for the production of the trisaccharide kestose from sucrose. The isolated complementing cosmid termed p21R1 was mobilized into the parental strain by conjugal mating to increase the gene copy number and thereby the levansucrase yield, finally resulting in 189 units mL-1 representing more than 95% of the total proteins secreted under selected culture conditions (Hernandez et al. 1999). In a further study, this levansucrase and its D309N derivative were enzymatically characterized after cloning their genes into broad-host-range cloning vector pRK293 and expression from the native lsdA promoter (Batista et al. 1999). A later study demonstrated the importance of lsdA for adaptation to high osmotic stress and for biofilm formation (Velazquez-Hernandez et al. 2011). The lsdA mutant showed a decreased tolerance to 50–150 mM NaCl, a 99% reduced tolerance to desiccation, and a decrease in the ability to form cell aggregates on abiotic surfaces. Complementation of the mutant by expressing lsdA again from its native promoter, both cloned as a fragment into broad-host-range plasmid pBBR1MCS-3 (see Gluconobacter section), recovered the abilities of the parental strain in the lsdA disruptant mutant.
Ga. diazotrophicus as an endophytic nitrogen-fixing plant-associated microbe is generally also interesting as a host to express genes encoding proteins that could enable control of pests attacking agricultural plants. In this context, the Bacillus thuringiensis cry genes are interesting since they encode δ-endotoxins exhibiting entomopathogenic activity. To test the control of coleopteran pests in sugarcane, the cry3A gene hooked in the transposon Tn5 was introduced into Ga. diazotrophicus by conjugation on the narrow-host-range suicide plasmid backbone pSUP1021 carrying the transfer origin oriT of plasmid RP4 and integrating into the genome (Salles et al. 2000). The cry3A gene was successfully expressed in nitrogen-fixing conditions when fused to the promoter of the nifHDK operon from Rhizobium leguminosarum biovar trifolii. The use of regulated promoters activated only under specific conditions may avoid the problem of insect resistance caused by constant production of the δ-endotoxin. Later, the Cry1Ac gene from B. thuringiensis var. kurstaki was introduced into Ga. diazotrophicus after cloning into the double replicon shuttle vector pKT230 (Subashini et al. 2011). Clones containing the Cry1Ac gene produced the 130 kDa toxin protein both when present inside the sugarcane tissue as well as ex planta, and it did not impair the process of nitrogen fixation. To evaluate the colonization process of sugarcane plantlets and hydroponically grown rice seedlings by Ga. diazotrophicus, strain PAL5 was marked with gusA encoding β-glucuronidase and gfp encoding green fluorescent protein (Rouws et al. 2010). The gusA and gfp reporter genes were constitutively expressed under the control of the gentamicin resistance gene promoter on the broad-host-range plasmid backbone pBBR1. The reporter plasmids were shown to be valid tools to detect strain PAL5 and monitor colonization.
Recently, pBBR1MCS-5 (see Gluconobacter section) containing the mCherry gene under control of the promoter Ptac was used to analyze the colonization of Ga. diazotrophicus on two genotypes of elephant grass (Pennisetum purpureum), a perennial C4-plant employed for grazing, silage production or bioenergy, via seed coat and leaf spray (Camelo et al. 2020). The results showed that one elephant grass genotype produced a higher amount of biomass and total nitrogen than the other when Ga. diazotrophicus was sprayed on leaves. When colonizing plants and performing nitrogen fixation, the limited availability of iron might impose restrictions on the expression, assembly, and function of the nitrogenase complex. To better understand the iron uptake mechanisms in Ga. diazotrophicus, an ATP-binding cassette (ABC) transport system comprising the three genes feuABC encoding a periplasmic-binding protein, a permease, and a traffic ATPase was analyzed earlier (Urzua et al. 2013). In this study, the feuABC operon was expressed from its native promoter on a pJB3Tc20-based plasmid backbone with the RK2 replicon in a Ga. diazotrophicus parental strain and its feuAB disruptant mutant under iron-replete and iron-depleted conditions. The complementation of the mutant with the plasmid carrying the feuABC operon restored growth under iron-restricted conditions to the level seen for the parental strain, indicating that feuABC is needed for iron uptake.
Promoter screening in Ga. diazotrophicus
For Ga. diazotrophicus, a promoter library was constructed using the promoter probe vector pPW452, a derivative of the broad-host-range vector pMP220 obtained from pTJS75 with an RK2 replicon and containing a multiple cloning site upstream of a promoterless lacZ reporter gene (Schwab et al. 2016). With the library prepared from total Ga. diazotrophicus DNA, 480 clones were screened and six promoters were isolated and characterized further. They showed variable expression strengths in the presence of organic acids and polyalcohols, or glucose and sugarcane juices. The characterized promoters might be used in the future for the expression of genes of interest in Ga. diazotrophicus. Interestingly, three out of the six characterized promoters are probably involved in the transcription of antisense RNA, including the one showing the highest expression strength. However, a tight and strongly conditionally regulated promoter was not revealed in this study.
Target gene expression in Acidomonas
As described before, the AAB genus Acidomonas was established since several characteristics of the former species Acetobacter methanolicus could be distinguished from other type and representative AAB including Acetobacter strains, resulting in the renaming of A. methanolicus to Acidomonas methanolica (Urakami et al. 1989). For Acidomonas expression vectors including a basic promoter probe vector have been created based on the popular broad-host-range multi-copy plasmid RSF1010 (Schröder et al. 1989; Schröder et al. 1991). In A. methanolica, a RSF1010-based derivative was used to express the core antigen gene of hepatitis B virus subtype ayw under control of the E. coli Plac promoter alone and under tandem control of the strong promoter Pacm from the Acetobacter phage Acml and from Plac. When produced from the tandem promoters, the core antigen detected in lysates of A. methanolica was about 7-fold higher than in E. coli (Schröder et al. 1991).
The Pacm promoter and the E. coli promoters Plac and the temperature-dependent phage lambda PR promoter were tested in a RSF1010-based vector for expression of the pectate lyase gene ptlB from Erwinia carotovora and the aspartokinase/homoserine dehydrogenase genes thrAB from E. coli in A. methanolica (Föllner et al. 1994). The highest specific activities of the enzymes were detected with PR in tandem arrangement with ptlB and its native promoter and with Pacm in tandem with thrAB and its native promoter. A temperature shift to activate the Pr promoter was not needed in A. methanolica and was even disadvantageous, since it drastically lowered the resulting specific enzyme activity. The created vector system was also stable in A. methanolica in antibiotics-free media.
The RSF1010-based promoter probe vector pRS201 carrying a promoterless lacZ gene as a reporter was used to analyze the induction of the stationary growth phase promoters PbolAP1 and Pfic from E. coli in Gram-negative bacteria including A. methanolica (Miksch and Dobrowolski 1995). Transcriptional activation of Pfic in A. methanolica was growth phase-dependent as in E. coli and was increased 6- to 7-fold during the transition to the stationary phase. Subsequently, Pfic was used to test growth phase-dependent expression and production of a secreted hybrid β-glucanase consisting of 107 amino-terminal residues of Bacillus amyloliquefaciens β-glucanase and 107 carboxy-terminal residues of B. macerans β-glucanase in A. methanolica using a Tn5-based transposon cassette non-specifically integrating into the chromosome (Miksch et al. 1997). Due to Pfic, the β-glucanase was highly overexpressed and secreted into the medium during the stationary phase, while total and extracellular production of β-glucanase varied depending on the transposon integration site. The viability of the bacterial cells was not affected and cell lysis did not occur.
Target gene expression in Acidiphilium
The transfer of genetic information into Acidiphilium and the construction of suitable vectors started about 30 years ago (Roberto et al. 1991). Plasmids from different incompatibility groups have been tested to determine the frequency with which they are transferred from E. coli to Acidiphilium strains. During these studies, a conjugative function has been discovered in Acidiphilium strains by observing the transfer of mobilizable, but not self-transmissible plasmids between Acidiphilium cells (Roberto et al. 1991). In following studies, DNA transfer into Acidiphilium by conjugation but also by electroporation was established and improved (Glenn et al. 1992; Inagaki et al. 1993). The pRK2 replicon-based broad-host-range plasmids pRK415 and pLAFR3 were used to analyze transformation efficiencies, which turned out to depend on the recipient strains (Glenn et al. 1992). Also, the E. coli / Acidiphilium shuttle vector pAH101 was constructed by fusing a restriction fragment from the native pAH1 plasmid from Acidiphilium sp. 42H with a fragment carrying the β-lactamase gene from pUC19 as resistance marker. Transformants of A. facilis could grow in ampicillin-containing medium only with pAH101, showing that the β-lactamase gene was efficiently expressed from the native Tn3 promoter region also in A. facilis (Inagaki et al. 1993). However, the pAH101 plasmid appeared to be unstable in A. facilis in the stationary phase.
In further work, the plasmid pRK415 conferring tetracycline resistance was used to express the arsenic resistance genes arsABC from a natural E. coli isolate in Acidiphilium (Bruhn and Roberto 1993). After conjugal transfer of the constructed plasmid pIRC107 from E. coli to Acidiphilium, the plasmid was stable for at least one week with and without tetracycline selection. Also, the transformed Acidiphilium strain survived longer and in higher numbers in cultures with arsenopyrite ore than the parental strain did, suggesting a functional expression of the arsABC operon from its native E. coli promoter. Genetically improved strains of acidophilic bacteria were considered to have the potential to optimize microbial leaching of metals from ores. Enhancing their resistance to toxic metals is one option for strain improvement.
Acidiphilium could also be used to degrade organic pollutants in acidic wastewaters, provided they harbor the required genes. To evaluate the potential of Acidiphilium for biodegradation in acidic environments, gene transfer systems were tested by using the IncP1 antibiotic resistance plasmids RP4 and pVK101 and the phenol degradation-encoding plasmid pPGH11 (Quentmeier and Friedrich 1994). The plasmids RP4 from Pseudomonas, pVK101 (RK2 and pHK17, see Gluconobacter section for pVK) and pPGH11 (R68.45 and pPGH1 from Pseudomonas) could be transferred into A. cryptum with frequencies of 1.8 x 10-2 to 9.8 x 10-4 transconjugants per recipient cell. In the transconjugants, the antibiotic resistances and the ability to degrade phenol were expressed. A. cryptum with pPGH11 grew with 2.5 mM phenol at a doubling time of 12 h and a yield of 0.52 g dry cell weight per g of phenol.
In a later study, the native pAM5 plasmid from A. multivorum was sequenced and analyzed to use its characteristics for the construction of a shuttle vector (Singh and Banerjee 2007). The plasmid pSK2 containing the pAM5 rep region, the pBR322 ori and the ampicillin resistance gene ampR from pUC19, and a multiple cloning site, was able to replicate and stably maintained in E. coli, Acidiphilium as well as in Acidocella. The plasmid copy number of pAM5 and pSK2 in A. multivorum was determined to be 50-60. Apparently, no further study was published yet demonstrating the functional expression of target genes in Acidiphilium strains with pSK2.
Target gene expression in Asaia
Representatives from AAB genera have been demonstrated to be naturally associated not only with plants (e.g., Acetobacter, Asaia, Gluconacetobacter, Gluconobacter), but also with insects. The presence of AAB genera in insects (e.g., the fruit fly Drosophila melanogaster and the honeybee Apis mellifera), that typically use plant materials and sugar-rich matrices as food sources, is seen as roles of these AAB in exploitation of the food. Asaia was also found in the malaria mosquito vector Anopheles stephensi and is also present in and cross-colonizing other sugar-feeding insects of phylogenetically distant genera and orders (Crotti et al. 2009; Favia et al. 2007). In these studies, for tracking one Asaia strain was constructed by tagging it with a plasmid derivative of pHM2 carrying the pBR322 ori. The plasmid pHM2-Gfp was used to functionally express the green fluorescent protein Gfp under control of the neomycin phosphotransferase promoter Pnpt II found to be active in a broader range of species. Cross-colonization patterns of the body of the mosquitos An. stephensi, Aedes aegypti and the leafhopper Scaphoideus titanus have been documented with Asaia strains labeled with a chromosome- and a plasmid-encoded fluorescent protein. Another Asaia strain was constructed by the insertion of a mini-Tn5 gene cassette into the chromosome. This cassette contained the dsRed gene encoding a red fluorescent protein under control of the ribosomal promoter PrrnBP1 from E. coli. Fluorescence and confocal microscopy analysis showed that the labeled Asaia strains efficiently colonized guts, male and female reproductive systems and the salivary glands.
The ability to cross-colonize insects of phylogenetically distant orders is an important property for the development of symbiont-based control of different vector-borne diseases. To test the potential of such a control by activation of insect immunity, the pHM2 backbone was used in Asaia to express the Wolbachia surface protein (WSP) gene with its signal peptide sequence under the control of Pnpt II that was derived from a Wolbachia strain isolated from the nematode Dirofilaria immitis (Epis et al. 2020). The recombinant strain AsaiaWSP induced the activation of the host immune response in An. stephensi and Ae. aegypti mosquitos and it inhibited the development of the heartworm parasite D. immitis in Ae. aegypti. The recombinant AsaiaWSP was also found to activate macrophages and Leishmania killing stronger than the empty vector control strain did (Varotto-Boccazzi et al. 2020). These results consolidated the immune-stimulating property of WSP and make AsaiaWSP worth of further investigations as a potential tool for the control of mosquito-borne diseases and for immune prophylaxis and therapy of leishmaniases and other diseases that could be inhibited by macrophage activation.
Outlook
For AAB a diversity of expression plasmids has been developed and tested for expression of target genes and major plasmid lineages can be identified that are applied most often in AAB, although hitherto in only seven out of 49 AAB genera. Expression of target genes was achieved using the corresponding native promoter region or using a limited number of characterized promoters. Apparently, Plac allowing LacI-dependent induction of expression in E. coli is the most used heterologous promoter for expression in AAB. Varying the length of the Plac region can increase or decrease target gene expression in Acetobacter likely due to 5’-UTR effects in the mRNA (Tonouchi et al. 1998a). Only two studies reported to have used LacI-dependent IPTG-induced target gene expression (Bugala et al. 2016; Liu et al. 2020b). Only in one study the LacI-dependent IPTG-induced expression performance has been reported (Liu et al. 2020b). By using the fluorescence reporter gene mRFP1, the results showed that according to fluorescence microscope analysis, only a fraction of K. xylinus cells showed mRFP1 fluorescence; thus, cell heterogeneity was observed. Furthermore, only 3-fold induction by IPTG was observed from 0.1 mM to 0.5 mM reaching a saturation level of mRFP1 signals at 0.4 mM and 0.5 mM, while the 0 mM condition was not reported yet. Thus, the performance of IPTG-induced expression in AAB still remains to be solved or at least to be improved for tight and homogenous LacI-dependent target gene expression.
Due to a deficiency in regulatable promoters, almost only constitutive promoters have been used in AAB. The greatest variety of endogenous, heterologous, and synthetic promoters functional in AAB and exhibiting weak, moderate, strong, or very strong activity is known for Gluconobacter and Komagataeibacter (Florea et al. 2016a; Hölscher and Görisch 2006; Hu et al. 2015; Kallnik et al. 2010; Mientus et al. 2017; Shi et al. 2014; Teh et al. 2019; Yuan et al. 2016a). These data will be helpful to test and broaden the range of usable promoters in other AAB genera. For Gluconobacter, the highest change of a promoter activity according to plasmid-based LacZ activities was reported for the promoter Pidh of the inositol dehydrogenase gene GOX1857 from G. oxydans 621H (Mientus et al. 2017). Depending on the carbon source, Pidh activity was very low on mannitol and even lower on glucose, while Pidh activity on sorbitol was 100-fold higher than on glucose. This probably is the highest fold-change difference reported so far for Gluconobacter or AAB in general. However, the molecular mechanism of Pidh regulation and the transcriptional regulator(s) of Pidh involved need to be unraveled as well as the performance and the suitability of Pidh for use in a regulatable endogenous expression system. The R. leguminosarum promoter of the nifHDK operon, which is activated under nitrogen-fixing conditions, was functional in plant-associated Ga. diazotrophicus in planta. Therefore, PnifHDK might also be used in nitrogen-fixing AAB ex planta to control gene expression in dependence of the available nitrogen sources (Salles et al. 2000). The basal expression (tightness), induction performance and relative induced expression strength of the heterologous PnifHDK promoter applied in AAB were not reported yet. Likewise, endogenous nif promoters from nitrogen-fixing AAB strains could be characterized and, if suitable, used to construct regulatable expression systems in nitrogen-fixing AAB. The system(s) could possibly be transferred with all regulatory requirements to non-nitrogen-fixing AAB.
Until now, for AAB only two well-documented inducible heterologous expression systems are available for only two species from different genera. As described above, one is based on an AHL-inducible luxR-Plux system and the other one is a well-tunable l-arabinose-inducible araC-ParaBAD system with up to 480-fold induction (Florea et al. 2016a; Fricke et al. 2020). The AHL-inducible luxR-Plux system was reported to enable strong induction in Komagataeibacter, yet the strength of the induction appeared to depend on the growth conditions. Induction ranged from only up to 5-fold due to high leakiness in the absence of the inducer to a very much better induction performance in cells inside cellulose pellicles (Florea et al. 2016a). Understanding the reason for this difference in induction performance may help to further improve the luxR-Plux system and possibly other heterologous expression systems (e.g., TetR) still leaky in Komagataeibacter for full functionality in any growth condition. In Komagataeibacter, an l-arabinose-inducible araC-ParaBAD system was also very leaky and thus induction fold-change was very low (Teh et al. 2019). In contrast, in G. oxydans 621H, the heterologous araC-ParaBAD system from E. coli MC4100 was very tight and tunable by l-arabinose concentrations up to 1% (w/v) (Fricke et al. 2020). However, in G. oxydans 621H, the l-arabinose oxidation activity is high and contributes to the strong acidification of the growth medium by forming arabinonic acid causing a severe loss of reporter activities, while the d-arabinose oxidation activity is low (Fricke et al. 2020; Mientus et al. 2017; Peters et al. 2017). Thus, using d-arabinose-responsive AraC derivatives could circumvent the pH issue of l-arabinose induction and might probably also result in increased sensitivity toward arabinose concentrations. Therefore, engineered AraC mutant proteins with altered binding pockets which activate transcription in response to d-arabinose and not in response to l-arabinose could be tested (Tang et al. 2008). The AraC protein was also engineered to specifically respond to triacetic acid lactone, vanillin and salicylic acid (Frei et al. 2018). The new phenolic-sensing variants of AraC showed responses of more than 100-fold over the background in E. coli and were highly specific toward their target compound. That illustrates the potential of the AraC transcriptional regulatory protein for molecular sensing, reporting and target gene expression, which could possibly also be achieved and used in AAB.
In summary, after 35 years of constitutive target gene expression in AAB, we now have the first regulatable expression systems for AAB in hand. Further promising candidates are in sight for both heterologous and endogenous regulatable expression systems and future studies certainly will reveal more regulated AAB promoters. Additionally, the flexible SEVA toolkit, used within the AAB hitherto only in Komagataeibacter, is expected to speed up expression system development and testing or screening of such systems in even more AAB genera.
References
Akasaka N, Astuti W, Ishii Y, Hidese R, Sakoda H, Fujiwara S (2015) Change in the plasmid copy number in acetic acid bacteria in response to growth phase and acetic acid concentration. J Biosci Bioeng 119(6):661–668. https://doi.org/10.1016/j.jbiosc.2014.11.003
Antoine R, Locht C (1992) Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol Microbiol 6(13):1785–1799. https://doi.org/10.1111/j.1365-2958.1992.tb01351.x
Arrieta J, Hernandez L, Coego A, Suarez V, Balmori E, Menendez C, PetitGlatron MF, Chambert R, SelmanHousein G (1996) Molecular characterization of the levansucrase gene from the endophytic sugarcane bacterium Acetobacter diazotrophicus SRT4. Microbiol 142:1077–1085. https://doi.org/10.1099/13500872-142-5-1077
Batista FR, Hernandez L, Fernandez JR, Arrieta J, Menendez C, Gomez R, Tambara Y, Pons T (1999) Substitution of Asp-309 by Asn in the Arg-Asp-Pro (RDP) motif of Acetobacter diazotrophicus levansucrase affects sucrose hydrolysis, but not enzyme specificity. Biochem J 337:503–506. https://doi.org/10.1042/0264-6021:3370503
Battad-Bernardo E, McCrindle SL, Couperwhite I, Neilan BA (2004) Insertion of an E. coli lacZ gene in Acetobacter xylinus for the production of cellulose in whey. FEMS Microbiol Lett 231(2):253–260. https://doi.org/10.1016/S0378-1097(04)00007-2
Battling S, Wohlers K, Igwe C, Kranz A, Pesch M, Wirtz A, Baumgart M, Büchs J, Bott M (2020) Novel plasmid-free Gluconobacter oxydans strains for production of the natural sweetener 5-ketofructose. Microbiol Cell Fact 19(1) ARTN 54. https://doi.org/10.1186/s12934-020-01310-7
Bilska V, Grones J (2003) Optimization of transformation and electroporation of plasmid DNA with pAC1 replicon into Acetobacter pasteurianus. Biologia 58(6):1029–1035
Blank M, Schweiger P (2018) Surface display for metabolic engineering of industrially important acetic acid bacteria. PeerJ 6. ARTN e4626. https://doi.org/10.7717/peerj.4626
Bruhn DF, Roberto FF (1993) Maintenance and expression of enteric arsenic resistance genes in Acidiphilium. Biohydrometallurgical Technologies 2:745–754
Bugala J, Cimova V, Grones P, Grones J (2016) Characterization of newly identified DnaA and DnaB proteins from Acetobacter. Res Microbiol 167(8):655–668. https://doi.org/10.1016/j.resmic.2016.06.010
Camelo A, Barreto CP, Vidal MS, Rouws JRC, Ledo FJD, Schwab S, Baldani JI (2020) Field response of two seed propagated elephant grass genotypes to diazotrophic bacterial inoculation and in situ confocal microscopy colonization analyses. Symbiosis. https://doi.org/10.1007/s13199-020-00730-8
Chen R (2012) Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnol Adv 30(5):1102–1107. https://doi.org/10.1016/j.biotechadv.2011.09.013
Chien LJ, Chen HT, Yang PF, Lee CK (2006) Enhancement of cellulose pellicle production by constitutively expressing Vitreoscilla hemoglobin in Acetobacter xylinum. Biotechnol Progr 22(6):1598–1603. https://doi.org/10.1021/bp060157g
Cleenwerck I, De Vos P (2008) Polyphasic taxonomy of acetic acid bacteria: An overview of the currently applied methodology. Int J Food Microbiol 125(1):2–14. https://doi.org/10.1016/j.ijfoodmicro.2007.04.017
Cleton-Jansen AM, Dekker S, Vandeputte P, Goosen N (1991) A single amino acid substitution changes the substrate specificity of quinoprotein glucose dehydrogenase in Gluconobacter oxydans. Mol Gen Genet 229(2):206–212. https://doi.org/10.1007/Bf00272157
Cohen SN, Chang ACY, Boyer HW, Helling RB (1973) Construction of biologically functional bacterial plasmids in vitro. P Natl Acad Sci USA 70(11):3240–3244. https://doi.org/10.1073/pnas.70.11.3240
Condon C, Fitzgerald RJ, Ogara F (1991) Conjugation and heterologous gene expression in Gluconobacter oxydans ssp suboxydans. FEMS Microbiol Lett 80(2-3):173–178. https://doi.org/10.1016/0378-1097(91)90590-7
Connell ND (2001) Expression systems for use in actinomycetes and related organisms. Curr Opin Biotechnol 12(5):446–449. https://doi.org/10.1016/S0958-1669(00)00243-3
Creaven M, Fitzgerald RJ, Ogara F (1994) Transformation of Gluconobacter oxydans subsp. suboxydans by electroporation. Can J Microbiol 40(6):491–494. https://doi.org/10.1139/m94-079
Crotti E, Damiani C, Pajoro M, Gonella E, Rizzi A, Ricci I, Negri I, Scuppa P, Rossi P, Ballarini P, Raddadi N, Marzorati M, Sacchi L, Clementi E, Genchi M, Mandrioli M, Bandi C, Favia G, Alma A, Daffonchio D (2009) Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ Microbiol 11(12):3252–3264. https://doi.org/10.1111/j.1462-2920.2009.02048.x
De Roos J, De Vuyst L (2018) Acetic acid bacteria in fermented foods and beverages. Curr Opin Biotechnol 49:115–119. https://doi.org/10.1016/j.copbio.2017.08.007
Deeraksa A, Moonmangmee S, Toyama H, Adachi O, Matsushita K (2006) Conversion of capsular polysaccharide, involved in pellicle formation, to extracellular polysaccharide by galE deletion in Acetobacter tropicalis. Biosci Biotechnol Biochem 70(10):2536–2539. https://doi.org/10.1271/bbb.60143
Deng Y, Nagachar N, Fang L, Luan X, Catchmark JM, Tien M, Kao TH (2015) Isolation and characterization of two cellulose morphology mutants of Gluconacetobacter hansenii ATCC23769 producing cellulose with lower crystallinity. PLOS One 10(3) ARTN e0119504. https://doi.org/10.1371/journal.pone.0119504
Deng Y, Nagachar N, Xiao CW, Tien M, Kao TH (2013) Identification and characterization of non-cellulose-producing mutants of Gluconacetobacter hansenii generated by Tn5 transposon mutagenesis. J Bacteriol 195(22):5072–5083. https://doi.org/10.1128/Jb.00767-13
Dilworth MV, Piel MS, Bettaney KE, Ma P, Luo J, Sharples D, Poyner DR, Gross SR, Moncoq K, Henderson PJF, Miroux B, Bill RM (2018) Microbial expression systems for membrane proteins. Methods 147:3–39. https://doi.org/10.1016/j.ymeth.2018.04.009
Durante-Rodriguez G, de Lorenzo V, Martinez-Garcia E (2014) The Standard European Vector Architecture (SEVA) plasmid toolkit. Pseudomonas: Methods and Protocols 1149:469–478. https://doi.org/10.1007/978-1-4939-0473-0_36
Epis S, Varotto-Boccazzi I, Crotti E, Damiani C, Giovati L, Mandrioli M, Biggiogera M, Gabrieli P, Genchi M, Polonelli L, Daffonchio D, Favia G, Bandi C (2020) Chimeric symbionts expressing a Wolbachia protein stimulate mosquito immunity and inhibit filarial parasite development. Commun Biol 3(1) ARTN 105. https://doi.org/10.1038/s42003-020-0835-2
Evans JC, Mizrahi V (2015) The application of tetracyclineregulated gene expression systems in the validation of novel drug targets in Mycobacterium tuberculosis. Front Microbiol 6:812. https://doi.org/10.3389/fmicb.2015.00812
Fang J, Kawano S, Tajima K, Kondo T (2015) In vivo curdlan/cellulose bionanocomposite synthesis by genetically modified Gluconacetobacter xylinus. Biomacromolecules 16(10):3154–3160. https://doi.org/10.1021/acs.biomac.5b01075
Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, Rizzi A, Urso R, Brusetti L, Borin S, Mora D, Scuppa P, Pasqualini L, Clementi E, Genchi M, Corona S, Negri I, Grandi G, Alma A, Kramer L, Esposito F, Bandi C, Sacchi L, Daffonchio D (2007) Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. P Natl Acad Sci USA 104(21):9047–9051. https://doi.org/10.1073/pnas.0610451104
Felder M, Gupta A, Verma V, Kumar A, Qazi GN, Callum J (2000) The pyrroloquinoline quinone synthesis genes of Gluconobacter oxydans. FEMS Microbiol Lett 193(2):231–236. https://doi.org/10.1016/S0378-1097(00)00487-0
Fjaervik E, Frydenlund K, Valla S, Huggirat Y, Benziman M (1991) Complementation of cellulose-negative mutants of Acetobacter xylinum by the cloned structural gene for phosphoglucomutase. FEMS Microbiol Lett 77(2-3):325–330. https://doi.org/10.1111/j.1574-6968.1991.tb04370.x
Florea M, Hagemann H, Santosa G, Abbott J, Micklem CN, Spencer-Milnes X, Garcia LD, Paschou D, Lazenbatt C, Kong DZ, Chughtai H, Jensen K, Freemont PS, Kitney R, Reeve B, Ellis T (2016a) Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. P Natl Acad Sci USA 113(24):E3431–E3440. https://doi.org/10.1073/pnas.1522985113
Florea M, Reeve B, Abbott J, Freemont PS, Ellis T (2016b) Genome sequence and plasmid transformation of the model high-yield bacterial cellulose producer Gluconacetobacter hansenii ATCC 53582. Sci Rep 6. ARTN 23635. https://doi.org/10.1038/srep23635
Föllner CG, Schröder R, Babel W (1994) Construction of broad-host-range plasmids for the expression of heterologous genes in Acetobacter methanolicus B58. Acta Biotechnol 14(2):141–151. https://doi.org/10.1002/abio.370140205
Forstner M, Leder L, Mayr LM (2007) Optimization of protein expression systems for modern drug discovery. Expert Rev Prot 4(1):67–78. https://doi.org/10.1586/14789450.4.1.67
Frei CS, Qian S, Cirino PC (2018) New engineered phenolic biosensors based on the AraC regulatory protein. Protein Eng Des Sel 31(6):213–220. https://doi.org/10.1093/protein/gzy024
Fricke PM, Link T, Gätgens J, Sonntag C, Otto M, Bott M, Polen T (2020) A tunable L-arabinose-inducible expression plasmid for the acetic acid bacterium Gluconobacter oxydans. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-020-10905-4
Fujiwara M, Fukushi K, Takai M, Hayashi J, Fukaya M, Okumura H, Kawamura Y (1992) Construction of shuttle vectors derived from Acetobacter xylinum for cellulose-producing bacterium Acetobacter xylinum. Biotechnol Lett 14(7):539–542. https://doi.org/10.1007/Bf01023936
Fukaya M, Okumura H, Masai H, Uozumi T, Beppu T (1985a) Construction of new shuttle vectors for Acetobacter. Agr Biol Chem 49(7):2083–2090. https://doi.org/10.1080/00021369.1985.10867038
Fukaya M, Okumura H, Masai H, Uozumi T, Beppu T (1985b) Development of a host-vector system for Gluconobacter suboxydans. Agr Biol Chem 49(8):2407–2411. https://doi.org/10.1080/00021369.1985.10867095
Fukaya M, Takemura H, Okumura H, Kawamura Y, Horinouchi S, Beppu T (1990) Cloning of genes responsible for acetic acid resistance in Acetobacter aceti. J Bacteriol 172(4):2096–2104. https://doi.org/10.1128/jb.172.4.2096-2104.1990
Fukaya M, Takemura H, Tayama K, Okumura H, Kawamura Y, Horinouchi S, Beppu T (1993) The aarC gene responsible for acetic acid assimilation confers acetic acid resistance on Acetobacter aceti. J Ferment Bioeng 76(4):270–275. https://doi.org/10.1016/0922-338x(93)90192-B
Fukaya M, Tayama K, Okumura H, Masai H, Uozumi T, Beppu T (1985c) Improved transformation method for Acetobacter with plasmid DNA. Agr Biol Chem 49(7):2091–2097. https://doi.org/10.1080/00021369.1985.10867039
Fukaya M, Tayama K, Tamaki T, Tagami H, Okumura H, Kawamura Y, Beppu T (1989) Cloning of the membrane-bound aldehyde dehydrogenase gene of Acetobacter polyoxogenes and improvement of acetic acid production by use of the cloned gene. Appl Environ Microbiol 55(1):171–176. https://doi.org/10.1128/Aem.55.1.171-176.1989
Gao L, Wu XD, Xia XL, Jin ZY (2020a) Fine-tuning ethanol oxidation pathway enzymes and cofactor PQQ coordinates the conflict between fitness and acetic acid production by Acetobacter pasteurianus. Microbiol Biotechnol. https://doi.org/10.1111/1751-7915.13703
Gao L, Wu XD, Zhu CL, Jin ZY, Wang W, Xia XL (2020b) Metabolic engineering to improve the biomanufacturing efficiency of acetic acid bacteria: advances and prospects. Crit Rev Biotechnol 40(4):522–538. https://doi.org/10.1080/07388551.2020.1743231
Gao LL, Hu YD, Liu J, Du GC, Zhou JW, Chen J (2014) Stepwise metabolic engineering of Gluconobacter oxydans WSH-003 for the direct production of 2-keto-L-gulonic acid from D-sorbitol. Metab Eng 24:30–37. https://doi.org/10.1016/j.ymben.2014.04.003
Gao LL, Liu YF, Zhang XY, Zhang HS (2020c) Efficient optimization of Gluconobacter oxydans based on protein scaffold-trimeric CutA to enhance the chemical structure stability of enzymes for the direct production of 2-keto-L-gulonic acid. J Chem 2020. ARTN 5429409. https://doi.org/10.1155/2020/5429409
Gätgens C, Degner U, Bringer-Meyer S, Herrmann U (2007) Biotransformation of glycerol to dihydroxyacetone by recombinant Gluconobacter oxydans DSM 2343. Appl Microbiol Biotechnol 76(3):553–559. https://doi.org/10.1007/s00253-007-1003-z
Glenn AW, Roberto FF, Ward TE (1992) Transformation of Acidiphilium by electroporation and conjugation. Can J Microbiol 38(5):387–393. https://doi.org/10.1139/m92-065
Gomes RJ, Borges MD, Rosa MD, Castro-Gomez RJH, Spinosa WA (2018) Acetic acid bacteria in the food Industry: Systematics, characteristics and applications. Food Technol Biotechnol 56(2):139–151. https://doi.org/10.17113/ftb.56.02.18.5593
Grones J, Bencova K (1994) Cloning, Production and Secretion of Beta-Galactosidase in Acetobacter-Pasteurianus. Folia Microbiol 39(2):99–104. https://doi.org/10.1007/Bf02906802
Grones J, Turna J (1992) Construction of shuttle vectors for cloning in Escherichia coli and Acetobacter pasteurianus. Folia Microbiol 37(6):395–400. https://doi.org/10.1007/Bf02899895
Grones J, Turna J (1995) Transformation of microorganisms with the plasmid vector with the replicon from pAC1 from Acetobacter pasteurianus. Biochem Bioph Res Co 206(3):942–947. https://doi.org/10.1006/bbrc.1995.1133
Gruber S, Schwab H, Koefinger P (2015) Versatile plasmid-based expression systems for Gram-negative bacteria - General essentials exemplified with the bacterium Ralstonia eutropha H16. New Biotechnol 32(6):552–558. https://doi.org/10.1016/j.nbt.2015.03.015
Gullo M, La China S, Falcone PM, Giudici P (2018) Biotechnological production of cellulose by acetic acid bacteria: Current state and perspectives. Appl Microbiol Biotechnol 102(16):6885–6898. https://doi.org/10.1007/s00253-018-9164-5
Hall PE, Anderson SM, Johnston DM, Cannon RE (1992) Transformation of Acetobacter xylinum with plasmid DNA by electroporation. Plasmid 28(3):194–200. https://doi.org/10.1016/0147-619x(92)90051-B
Hernandez L, Ramirez R, Hormaza JV, Madrazo J, Arrieta J (1999) Increased levansucrase production by a genetically modified Acetobacter diazotrophicus strain in shaking batch cultures. Lett Appl Microbiol 28(1):41–44. https://doi.org/10.1046/j.1365-2672.1999.00470.x
Herweg E, Schopping M, Rohr K, Siemen A, Frank O, Hofmann T, Deppenmeier U, Büchs J (2018) Production of the potential sweetener 5-ketofructose from fructose in fed-batch cultivation with Gluconobacter oxydans. Bioresource Technol 259:164–172. https://doi.org/10.1016/j.biortech.2018.03.038
Hoffmann JJ, Hövels M, Kosciow K, Deppenmeier U (2020) Synthesis of the alternative sweetener 5-ketofructose from sucrose by fructose dehydrogenase and invertase producing Gluconobacter strains. J Biotechnol 307:164–174. https://doi.org/10.1016/j.jbiotec.2019.11.001
Hölscher T, Görisch H (2006) Knockout and overexpression of pyrroloquinoline quinone biosynthetic genes in Gluconobacter oxydans 621H. J Bacteriol 188(21):7668–7676. https://doi.org/10.1128/Jb.01009-06
Hölscher T, Weinert-Sepalage D, Görisch H (2007) Identification of membrane-bound quinoprotein inositol dehydrogenase in Gluconobacter oxydans ATCC 621H. Microbiol 153:499–506. https://doi.org/10.1099/mic.0.2006/002196-0
Hövels M, Kosciow K, Kniewel J, Jakob F, Deppenmeier U (2020) High yield production of levan-type fructans by Gluconobacter japonicus LMG 1417. Int J Biol Macromol 164:295–303. https://doi.org/10.1016/j.ijbiomac.2020.07.105
Hu SQ, Gao YG, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, Munekata M, Tanaka I, Yao M (2010) Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. P Natl Acad Sci USA 107(42):17957–17961. https://doi.org/10.1073/pnas.1000601107
Hu YD, Wan H, Li JG, Zhou JW (2015) Enhanced production of L-sorbose in an industrial Gluconobacter oxydans strain by identification of a strong promoter based on proteomics analysis. J Ind Microbiol Biotechnol 42(7):1039–1047. https://doi.org/10.1007/s10295-015-1624-7
Huang LH, Liu QJ, Sun XW, Li XJ, Liu M, Jia SR, Xie YY, Zhong C (2020) Tailoring bacterial cellulose structure through CRISPR interference-mediated downregulation of galU in Komagataeibacter xylinus CGMCC 2955. Biotechnol Bioeng 117(7):2165–2176. https://doi.org/10.1002/bit.27351
Hur DH, Choi WS, Kim TY, Lee SY, Park JH, Jeong KJ (2020) Enhanced production of bacterial cellulose in Komagataeibacter xylinus via tuning of biosynthesis genes with synthetic RBS. J Microbiol Biotechnol 30(9):1430–1435. https://doi.org/10.4014/jmb.2006.06026
Iida A, Ohnishi Y, Horinouchi S (2008a) Control of acetic acid fermentation by quorum sensing via N-acylhomoserine lactones in Gluconacetobacter intermedius. J Bacteriol 190(7):2546–2555. https://doi.org/10.1128/Jb.01698-07
Iida A, Ohnishi Y, Horinouchi S (2008b) An OmpA family protein, a target of the GinI/GinR quorum-sensing system in Gluconacetobacter intermedius, controls acetic acid fermentation. J Bacteriol 190(14):5009–5019. https://doi.org/10.1128/Jb.00378-08
Iida A, Ohnishi Y, Horinouchi S (2009) Identification and characterization of target genes of the GinI/GinR quorum-sensing system in Gluconacetobacter intermedius. Microbiol 155:3021–3032. https://doi.org/10.1099/mic.0.028613-0
Inagaki K, Tomono J, Kishimoto N, Tano T, Tanaka H (1993) Transformation of the acidophilic heterotroph Acidiphilium facilis by electroporation. Biosci Biotechnol Biochem 57(10):1770–1771. https://doi.org/10.1271/bbb.57.1770
Ishida T, Sugano Y, Shoda M (2002) Novel glycosyltransferase genes involved in the acetan biosynthesis of Acetobacter xylinum. Biochem Bioph Res Co 295(2):230–235. Pii S0006-291x(02)00663-0. https://doi.org/10.1016/S0006-291x(02)00663-0
Jacek P, Ryngajllo M, Bielecki S (2019) Structural changes of bacterial nanocellulose pellicles induced by genetic modification of Komagataeibacter hansenii ATCC 23769. Appl Microbiol Biotechnol 103(13):5339–5353. https://doi.org/10.1007/s00253-019-09846-4
Jacek P, Szustak M, Kubiak K, Gendaszewska-Darmach E, Ludwicka K, Bielecki S (2018) Scaffolds for chondrogenic cells cultivation prepared from bacterial cellulose with relaxed fibers structure induced genetically. Nanomaterials 8(12) ARTN 1066. https://doi.org/10.3390/nano8121066
Jang WD, Kim TY, Kim HU, Shim WY, Ryu JY, Park JH, Lee SY (2019) Genomic and metabolic analysis of Komagataeibacter xylinus DSM 2325 producing bacterial cellulose nanofiber. Biotechnol Bioeng 116(12):3372–3381. https://doi.org/10.1002/bit.27150
Kallnik V, Meyer M, Deppenmeier U, Schweiger P (2010) Construction of expression vectors for protein production in Gluconobacter oxydans. J Biotechnol 150(4):460–465. https://doi.org/10.1016/j.jbiotec.2010.10.069
Kashima Y, Iijima M, Nakano T, Tayama K, Koizumi W, Udaka S, Yanagida F (2000) Role of intracellular esterases in the production of esters by Acetobacter pasteurianus. J Biosci Bioeng 89(1):81–83. https://doi.org/10.1016/S1389-1723(00)88055-X
Kashima Y, Nakajima W, Nakano T, Tayama K, Koizumi Y, Udaka S, Yanagida F (1999) Cloning and characterization of ethanol-regulated esterase genes in Acetobacter pasteurianus. J Biosci Bioeng 87(1):19–27. https://doi.org/10.1016/S1389-1723(99)80003-6
Kataoka N, Matsutani M, Yakushi T, Matsushita K (2015) Efficient production of 2,5-diketo-D-gluconate via heterologous expression of 2-ketogluconate dehydrogenase in Gluconobacter japonicus. Appl Environ Microbiol 81(10):3552–3560. https://doi.org/10.1128/Aem.04176-14
Kawai S, Goda-Tsutsumi M, Yakushi T, Kano K, Matsushita K (2013) Heterologous overexpression and characterization of a flavoprotein-cytochrome c complex fructose dehydrogenase of Gluconobacter japonicus NBRC3260. Appl Environ Microbiol 79(5):1654–1660. https://doi.org/10.1128/Aem.03152-12
Ke X, Yu PH, Hu ZC, Chen L, Sun XQ, Zheng YG (2019) Synergistic improvement of PQQ-dependent D-sorbitol dehydrogenase activity from Gluconobacter oxydans for the biosynthesis of miglitol precursor 6-(N-hydroxyethyl)-amino-6-deoxy-alpha-L-sorbofuranose. J Biotechnol 300:55–62. https://doi.org/10.1016/j.jbiotec.2019.05.007
Kiefler I, Bringer S, Bott M (2015) SdhE-dependent formation of a functional Acetobacter pasteurianus succinate dehydrogenase in Gluconobacter oxydans-a first step toward a complete tricarboxylic acid cycle. Appl Microbiol Biotechnol 99(21):9147–9160. https://doi.org/10.1007/s00253-015-6972-8
Kiefler I, Bringer S, Bott M (2017) Metabolic engineering of Gluconobacter oxydans 621H for increased biomass yield. Appl Microbiol Biotechnol 101(13):5453–5467. https://doi.org/10.1007/s00253-017-8308-3
Kim TS, Hui G, Li JL, Kalia VC, Muthusamy K, Sohng JK, Kim IW, Lee JK (2019) Overcoming NADPH product inhibition improves D-sorbitol conversion to L-sorbose. Sci Rep 9. ARTN 815. https://doi.org/10.1038/s41598-018-37401-0
Kolesovs S, Semjonovs P (2020) Production of bacterial cellulose from whey-current state and prospects. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-020-10803-9
Kondo K, Beppu T, Horinouchi S (1995) Cloning, sequencing, and characterization of the gene encoding the smallest subunit of the three-component membrane-bound alcohol dehydrogenase from Acetobacter pasteurianus. J Bacteriol 177(17):5048–5055. https://doi.org/10.1128/jb.177.17.5048-5055.1995
Kondo K, Horinouchi S (1997) Characterization of the genes encoding the three-component membrane-bound alcohol dehydrogenase from Gluconobacter suboxydans and their expression in Acetobacter pasteurianus. Appl Environ Microbiol 63(3):1131–1138. https://doi.org/10.1128/Aem.63.3.1131-1138.1997
Konjanda P, Yakushi T, Matsushita K, Theeragool G (2019) Enhanced growth and ethanol oxidation by overexpressed caiA gene encoding acyl-CoA dehydrogenase in Komagataeibacter medellinensis NBRC 3288. Chiang Mai J Sci 46(2):196–206
Kosciow K, Domin C, Schweiger P, Deppenmeier U (2016) Extracellular targeting of an active endoxylanase by a TolB negative mutant of Gluconobacter oxydans. J Ind Microbiol Biotechnol 43(7):989–999. https://doi.org/10.1007/s10295-016-1770-6
Kosciow K, Zahid N, Schweiger P, Deppenmeier U (2014) Production of a periplasmic trehalase in Gluconobacter oxydans and growth on trehalose. J Biotechnol 189:27–35. https://doi.org/10.1016/j.jbiotec.2014.08.029
Kostner D, Luchterhand B, Junker A, Volland S, Daniel R, Büchs J, Liebl W, Ehrenreich A (2015) The consequence of an additional NADH dehydrogenase paralog on the growth of Gluconobacter oxydans DSM3504. Appl Microbiol Biotechnol 99(1):375–386. https://doi.org/10.1007/s00253-014-6069-9
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166(1):175–176. https://doi.org/10.1016/0378-1119(95)00584-1
Kovach ME, Phillips RW, Elzer PH, Roop RM 2nd, Peterson KM (1994) pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16(5):800–802
Kranz A, Busche T, Vogel A, Usadel B, Kalinowski J, Bott M, Polen T (2018) RNAseq analysis of α-proteobacterium Gluconobacter oxydans 621H. BMC Genomics 19. ARTN 24. https://doi.org/10.1186/s12864-017-4415-x
Kranz A, Vogel A, Degner U, Kiefler I, Bott M, Usadel B, Polen T (2017) High precision genome sequencing of engineered Gluconobacter oxydans 621H by combining long nanopore and short accurate Illumina reads. J Biotechnol 258:197–205. https://doi.org/10.1016/j.jbiotec.2017.04.016
Kretova M, Grones J (2006) Construction of shuttle vectors from pAP1 plasmid for cloning into Escherichia coli and Acetobacter strains. J Food Nutr Res 45(3):110–114
La China S, Zanichelli G, De Vero L, Gullo M (2018) Oxidative fermentations and exopolysaccharides production by acetic acid bacteria: a mini review. Biotechnol Lett 40(9-10):1289–1302. https://doi.org/10.1007/s10529-018-2591-7
Laureys D, Britton SJ, De Clippeleer J (2020) Kombucha tea fermentation: A review. J Am Soc Brew Chem 78(3):165–174. https://doi.org/10.1080/03610470.2020.1734150
Li KF, Mao XL, Liu L, Lin JP, Sun M, Wei DZ, Yang SL (2016a) Overexpression of membrane-bound gluconate-2-dehydrogenase to enhance the production of 2-keto-D-gluconic acid by Gluconobacter oxydans. Microbiol Cell Fact 15. ARTN 121. https://doi.org/10.1186/s12934-016-0521-8
Li MH, Wu JA, Lin JP, Wei DZ (2010a) Expression of Vitreoscilla hemoglobin enhances cell growth and dihydroxyacetone production in Gluconobacter oxydans. Curr Microbiol 61(5):370–375. https://doi.org/10.1007/s00284-010-9621-6
Li MH, Wu JA, Liu X, Lin JP, Wei DZ, Chen H (2010b) Enhanced production of dihydroxyacetone from glycerol by overexpression of glycerol dehydrogenase in an alcohol dehydrogenase-deficient mutant of Gluconobacter oxydans. Bioresource Technol 101(21):8294–8299. https://doi.org/10.1016/j.biortech.2010.05.065
Li S, Zhang JL, Xu H, Feng XH (2016b) Improved xylitol production from D-arabitol by enhancing the coenzyme regeneration efficiency of the pentose phosphate pathway in Gluconobacter oxydans. J Agr Food Chem 64(5):1144–1150. https://doi.org/10.1021/acs.jafc.5b05509
Liu D, Ke X, Hu ZC, Zheng YG (2020a) Combinational expression of D-sorbitol dehydrogenase and pyrroloquinoline quinone increases 6-(N-hydroxyethyl)-amino-6-deoxy-alpha-L-sorbofuranose production by Gluconobacter oxydans through cofactor manipulation. Enzyme Microbiol Tech 141. ARTN 109670. https://doi.org/10.1016/j.enzmictec.2020.109670
Liu LP, Yang X, Zhao XJ, Zhang KY, Li WC, Xie YY, Jia SR, Zhong C (2020b) A lambda Red and FLP/FRT-mediated site-specific recombination system in Komagataeibacter xylinus and its application to enhance the productivity of bacterial cellulose. ACS Synth Biol 9(11):3171–3180. https://doi.org/10.1021/acssynbio.0c00450
Liu M, Li SQ, Xie YZ, Jia SR, Hou Y, Zou Y, Zhong C (2018) Enhanced bacterial cellulose production by Gluconacetobacter xylinus via expression of Vitreoscilla hemoglobin and oxygen tension regulation. Appl Microbiol Biotechnol 102(3):1155–1165. https://doi.org/10.1007/s00253-017-8680-z
Lu LF, Wei LJ, Zhu K, Wei DZ, Hua Q (2012) Combining metabolic engineering and adaptive evolution to enhance the production of dihydroxyacetone from glycerol by Gluconobacter oxydans in a low-cost way. Bioresource Technol 117:317–324. https://doi.org/10.1016/j.biortech.2012.03.013
Lynch KM, Zannini E, Wilkinson S, Daenen L, Arendt EK (2019) Physiology of acetic acid bacteria and their role in vinegar and fermented beverages. Compr Rev Food Sci F 18(3):587–625. https://doi.org/10.1111/1541-4337.12440
Mamlouk D, Gullo M (2013) Acetic acid bacteria: Physiology and carbon sources oxidation. Indian J Microbiol 53(4):377–384. https://doi.org/10.1007/s12088-013-0414-z
Mangayil R, Rajala S, Pammo A, Sarlin E, Luo J, Santala V, Karp M, Tuukkanen S (2017) Engineering and characterization of bacterial nanocellulose films as low cost and flexible sensor material. ACS Appl Mater Inter 9(22):19048–19056. https://doi.org/10.1021/acsami.7b04927
Marx CJ, Lidstrom ME (2001) Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiol 147:2065–2075. https://doi.org/10.1099/00221287-147-8-2065
Masud U, Matsushita K, Theeragool G (2010) Cloning and functional analysis of adhS gene encoding quinoprotein alcohol dehydrogenase subunit III from Acetobacter pasteurianus SKU1108. Int J Food Microbiol 138(1-2):39–49. https://doi.org/10.1016/j.ijfoodmicro.2009.12.027
Matsumoto N, Hattori H, Matsutani M, Matayoshi C, Toyama H, Kataoka N, Yakushi T, Matsushita K (2018) A single-nucleotide insertion in a drug transporter gene induces a thermotolerance phenotype in Gluconobacter frateurii by increasing the NADPH/NADP+ ratio via metabolic change. Appl Environ Microbiol 84(10) ARTN e00354-18. https://doi.org/10.1128/AEM.00354-18
Matsushita K, Yakushi T, Toyama H, Shinagawa E, Adachi O (1996) Function of multiple heme c moieties in intramolecular electron transport and ubiquinone reduction in the quinohemoprotein alcohol dehydrogenase cytochrome c complex of Gluconobacter suboxydans. J Biol Chem 271(9):4850–4857
Matsutani M, Yakushi T (2018) Pyrroloquinoline quinone-dependent dehydrogenases of acetic acid bacteria. Appl Microbiol Biotechnol 102(22):9531–9540. https://doi.org/10.1007/s00253-018-9360-3
Mehta K, Pfeffer S, Brown RM (2015) Characterization of an acsD disruption mutant provides additional evidence for the hierarchical cell-directed self-assembly of cellulose in Gluconacetobacter xylinus. Cellulose 22(1):119–137. https://doi.org/10.1007/s10570-014-0521-y
Merfort M, Herrmann U, Bringer-Meyer S, Sahm H (2006a) High-yield 5-keto-D-gluconic acid formation is mediated by soluble and membrane-bound gluconate-5-dehydrogenases of Gluconobacter oxydans. Appl Microbiol Biotechnol 73(2):443–451. https://doi.org/10.1007/s00253-006-0467-6
Merfort M, Herrmann U, Ha SW, Elfari M, Bringer-Meyer S, Gorisch H, Sahm H (2006b) Modification of the membrane-bound glucose oxidation system in Gluconobacter oxydans significantly increases gluconate and 5-keto-D-gluconic acid accumulation. Biotechnol J 1(5):556–563. https://doi.org/10.1002/biot.200600032
Meyer M, Schweiger P, Deppenmeier U (2013) Effects of membrane-bound glucose dehydrogenase overproduction on the respiratory chain of Gluconobacter oxydans. Appl Microbiol Biotechnol 97(8):3457–3466. https://doi.org/10.1007/s00253-012-4265-z
Meyer M, Schweiger P, Deppenmeier U (2015) Succinic semialdehyde reductase Gox1801 from Gluconobacter oxydans in comparison to other succinic semialdehyde-reducing enzymes. Appl Microbiol Biotechnol 99(9):3929–3939. https://doi.org/10.1007/s00253-014-6191-8
Mientus M, Kostner D, Peters B, Liebl W, Ehrenreich A (2017) Characterization of membrane-bound dehydrogenases of Gluconobacter oxydans 621H using a new system for their functional expression. Appl Microbiol Biotechnol 101(8):3189–3200. https://doi.org/10.1007/s00253-016-8069-4
Miksch G, Dobrowolski P (1995) Growth phase-dependent induction of stationary phase promoters of Escherichia coli in different Gram-negative bacteria. J Bacteriol 177(18):5374–5378. https://doi.org/10.1128/jb.177.18.5374-5378.1995
Miksch G, Fiedler E, Dobrowolski P, Flaschel E (1997) Controlled secretion into the culture medium of a hybrid beta-glucanase by Acetobacter methanolicus mediated by the kil gene of Escherichia coli located on a Tn5-derived transposon. Appl Microbiol Biotechnol 47(5):530–536. https://doi.org/10.1007/s002530050968
Mogi T, Ano Y, Nakatsuka T, Toyama H, Muroi A, Miyoshi H, Migita CT, Ui H, Shiomi K, Omura S, Kita K, Matsushita K (2009) Biochemical and spectroscopic properties of cyanide-insensitive quinol oxidase from Gluconobacter oxydans. J Biochem 146(2):263–271. https://doi.org/10.1093/jb/mvp067
Mostafa HE, Heller KJ, Geis A (2002) Cloning of Escherichia coli lacZ and lacY genes and their expression in Gluconobacter oxydans and Acetobacter liquefaciens. Appl Environ Microbiol 68(5):2619–2623. https://doi.org/10.1128/Aem.68.5.2619-2623.2002
Nakai T, Moriya A, Tonouchi N, Tsuchida T, Yoshinaga F, Horinouchi S, Sone Y, Mori H, Sakai F, Hayashi T (1998) Control of expression by the cellulose synthase (bcsA) promoter region from Acetobacter xylinum BPR 2001. Gene 213(1-2):93–100. https://doi.org/10.1016/S0378-1119(98)00191-7
Nakai T, Nishiyama Y, Kuga S, Sugano Y, Shoda M (2002) ORF2 gene involves in the construction of high-order structure of bacterial cellulose. Biochem Bioph Res Co 295(2):458–462. Pii S0006-291x(02)00696-4. https://doi.org/10.1016/S0006-291x(02)00696-4
Nakai T, Sugano Y, Shoda M, Sakakibara H, Oiwa K, Tuzi S, Imai T, Sugiyama J, Takeuchi M, Yamauchi D, Mineyuki Y (2013) Formation of highly twisted ribbons in a carboxymethylcellulase gene-disrupted strain of a cellulose-producing bacterium. J Bacteriol 195(5):958–964. https://doi.org/10.1128/Jb.01473-12
Nakai T, Tonouchi N, Konishi T, Kojima Y, Tsuchida T, Yoshinaga F, Sakai F, Hayashi T (1999) Enhancement of cellulose production by expression of sucrose synthase in Acetobacter xylinum. P Natl Acad Sci USA 96(1):14–18. https://doi.org/10.1073/pnas.96.1.14
Nakano S, Fukaya M, Horinouchi S (2004) Enhanced expression of aconitase raises acetic acid resistance in Acetobacter aceti. FEMS Microbiol Lett 235(2):315–322. https://doi.org/10.1016/j.femsle.2004.05.007
Nishikura-Imamura S, Matsutani M, Insomphun C, Vangnai AS, Toyama H, Yakushi T, Abe T, Adachi O, Matsushita K (2014) Overexpression of a type II 3-dehydroquinate dehydratase enhances the biotransformation of quinate to 3-dehydroshikimate in Gluconobacter oxydans. Appl Microbiol Biotechnol 98(7):2955–2963. https://doi.org/10.1007/s00253-013-5439-z
Okamoto-Kainuma A, Yan W, Fukaya M, Tukamoto Y, Ishikawa M, Koizumi Y (2004) Cloning and characterization of the dnaKJ operon in Acetobacter aceti. J Biosci Bioeng 97(5):339–342. https://doi.org/10.1016/S1389-1723(04)70216-9
Okamoto-Kainuma A, Yan W, Kadono S, Tayama K, Koizumi Y, Yanagida F (2002) Cloning and characterization of groESL operon in Acetobacter aceti. J Biosci Bioeng 94(2):140–147. https://doi.org/10.1263/jbb.94.140
Okumura H, Tagami H, Fukaya M, Masai H, Kawamura Y, Horinouchi S, Beppu T (1988) Cloning of the β-isopropylmalate dehydrogenase gene from Acetobacter aceti and its use for construction of a new host-vector system for Acetobacter. Agr Biol Chem 52(12):3125–3129. https://doi.org/10.1080/00021369.1988.10869191
Okumura H, Uozumi T, Beppu T (1985) Construction of plasmid vectors and a genetic transformation system for Acetobacter aceti. Agr Biol Chem 49(4):1011–1017. https://doi.org/10.1080/00021369.1985.10866859
Parachin NS, Mulder KC, Viana AAB, Dias SC, Franco OL (2012) Expression systems for heterologous production of antimicrobial peptides. Peptides 38(2):446–456. https://doi.org/10.1016/j.peptides.2012.09.020
Peters B, Mientus M, Kostner D, Daniel R, Liebl W, Ehrenreich A (2017) Expression of membrane-bound dehydrogenases from a mother of vinegar metagenome in Gluconobacter oxydans. Appl Microbiol Biotechnol 101(21):7901–7912. https://doi.org/10.1007/s00253-017-8479-y
Petroni EA, Bocca SN, Ielpi L (1996) Sequence-specific DNA modification in Acetobacter xylinum. Cell Mol Biol 42(5):759–767
Phathanathavorn T, Naloka K, Matsutani M, Yakushi T, Matsushita K, Theeragool G (2019) Mutated fabG gene encoding oxidoreductase enhances the cost-effective fermentation of jasmine rice vinegar in the adapted strain of Acetobacter pasteurianus SKU1108. J Biosci Bioeng 127(6):690–697. https://doi.org/10.1016/j.jbiosc.2018.12.006
Prust C, Hoffmeister M, Liesegang H, Wiezer A, Fricke WF, Ehrenreich A, Gottschalk G, Deppenmeier U (2005) Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat Biotechnol 23(2):195–200. https://doi.org/10.1038/nbt1062
Quentmeier A, Friedrich CG (1994) Transfer and expression of degradative and antibiotic resistance plasmids in acidophilic bacteria. Appl Environ Microbiol 60(3):973–978. https://doi.org/10.1128/Aem.60.3.973-978.1994
Richhardt J, Bringer S, Bott M (2012) Mutational analysis of the pentose phosphate and Entner-Doudoroff pathways in Gluconobacter oxydans reveals improved growth of a Δedd Δeda mutant on mannitol. Appl Environ Microbiol 78(19):6975–6986. https://doi.org/10.1128/Aem.01166-12
Richhardt J, Luchterhand B, Bringer S, Büchs J, Bott M (2013) Evidence for a key role of cytochrome bo3 oxidase in respiratory energy metabolism of Gluconobacter oxydans. J Bacteriol 195(18):4210–4220. https://doi.org/10.1128/Jb.00470-13
Roberto FF, Glenn AW, Bulmer D, Ward TE (1991) Genetic transfer in acidophilic bacteria which are potentially applicable in coal beneficiation. Fuel 70(5):595–598. https://doi.org/10.1016/0016-2361(91)90172-7
Rouws LFM, Meneses CHSG, Guedes HV, Vidal MS, Baldani JI, Schwab S (2010) Monitoring the colonization of sugarcane and rice plants by the endophytic diazotrophic bacterium Gluconacetobacter diazotrophicus marked with gfp and gusA reporter genes. Lett Appl Microbiol 51(3):325–330. https://doi.org/10.1111/j.1472-765X.2010.02899.x
Saito Y, Ishii Y, Hayashi H, Imao Y, Akashi T, Yoshikawa K, Noguchi Y, Soeda S, Yoshida M, Niwa M, Hosoda J, Shimomura K (1997) Cloning of genes coding for L-sorbose and L-sorbosone dehydrogenases from Gluconobacter oxydans and microbial production of 2-keto-L-gulonate, a precursor of L-ascorbic acid, in a recombinant G. oxydans strain. Appl Environ Microbiol 63(2):454–460. https://doi.org/10.1128/Aem.63.2.454-460.1997
Saito Y, Ishii Y, Hayashi H, Yoshikawa K, Noguchi Y, Yoshida S, Soeda S, Yoshida M (1998) Direct fermentation of 2-keto-L-gulonic acid in recombinant Gluconobacter oxydans. Biotechnol Bioeng 58(2-3):309–315. https://doi.org/10.1002/(Sici)1097-0290(19980420)58:2/3<309::Aid-Bit30>3.3.Co;2-Q
Salles JF, Gitahy PD, Skot L, Baldani JI (2000) Use of endophytic diazotrophic bacteria as a vector to express the cry3A gene from Bacillus thuringiensis. Braz J Microbiol 31(3):155–161
Saxena IM, Kudlicka K, Okuda K, Brown RM (1994) Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum - Implications for cellulose crystallization. J Bacteriol 176(18):5735–5752. https://doi.org/10.1128/Jb.176.18.5735-5752.1994
Schleif R (2010) AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev 34(5):779–796. https://doi.org/10.1111/j.1574-6976.2010.00226.x
Schleyer U, Bringer-Meyer S, Sahm H (2008) An easy cloning and expression vector system for Gluconobacter oxydans. Int J Food Microbiol 125(1):91–95. https://doi.org/10.1016/j.ijfoodmicro.2007.04.016
Schnappinger D, Ehrt S (2014) Regulated expression systems for mycobacteria and their applications. Microbiol Spectr 2(1). https://doi.org/10.1128/microbiolspec.MGM2-0018-2013
Schröder R, Engel J, Chistoserdov AY, Tsygankov YD (1989) Construction of a promoter-probe vector for the methanol-utilizing bacterium Acetobacter methanolicus MB 58. Acta Biotechnol 9(3):219–225. https://doi.org/10.1002/abio.370090306
Schröder R, Maassen A, Lippoldt A, Borner T, Vonbaehr R, Dobrowolski P (1991) Expression of the core antigen gene of hepatitis B virus (Hbv) in Acetobacter methanolicus using broad-host-range vectors. Appl Microbiol Biotechnol 35(5):631–637. https://doi.org/10.1007/Bf00169628
Schwab S, Pessoa CA, Bergami AAD, Figueiredo NLD, Teixeira KRD, Baldani JI (2016) Isolation and characterization of active promoters from Gluconacetobacter diazotrophicus strain PAL5 using a promoter-trapping plasmid. Arch Microbiol 198(5):445–458. https://doi.org/10.1007/s00203-016-1203-y
Setyawati MI, Chien LJ, Lee CK (2007) Expressing Vitreoscilla hemoglobin in statically cultured Acetobacter xylinum with reduced O-2 tension maximizes bacterial cellulose pellicle production. J Biotechnol 132(1):38–43. https://doi.org/10.1016/j.jbiotec.2007.08.012
Setyawati MI, Chien LJ, Lee CK (2009) Self-immobilized recombinant Acetobacter xylinum for biotransformation. Biochem Eng J 43(1):78–84. https://doi.org/10.1016/j.bej.2008.09.002
Shen Y, Zhou X, Xu Y (2020) Enhancement of Gluconobacter oxydans resistance to lignocellulosic-derived inhibitors in xylonic acid production by overexpressing thioredoxin. Appl Biochem Biotechnol 191(3):1072–1083. https://doi.org/10.1007/s12010-020-03253-6
Shi LL, Li KF, Zhang H, Liu X, Lin JP, Wei DZ (2014) Identification of a novel promoter gHp0169 for gene expression in Gluconobacter oxydans. J Biotechnol 175:69–74. https://doi.org/10.1016/j.jbiotec.2014.01.035
Shi YY, Li KF, Lin JP, Yang SL, Wei DZ (2015) Engineered expression vectors significantly enhanced the production of 2-keto-D-gluconic acid by Gluconobacter oxidans. J Agr Food Chem 63(22):5492–5498. https://doi.org/10.1021/acs.jafc.5b01652
Shinjoh M, Hoshino T (1995) Development of a stable shuttle vector and a conjugative transfer system for Gluconobacter oxydans. J Ferment Bioeng 79(2):95–99. https://doi.org/10.1016/0922-338x(95)94074-2
Shinjoh M, Tomiyama N, Asakura A, Hoshino T (1995) Cloning and nucleotide sequencing of the membrane-bound L-sorbosone dehydrogenase gene of Acetobacter liquefaciens IFO 12258 and its expression in Gluconobacter oxydans. Appl Environ Microbiol 61(2):413–420. https://doi.org/10.1128/Aem.61.2.413-420.1995
Shinjoh M, Tomiyama N, Miyazaki T, Hoshino T (2002) Main polyol dehydrogenase of Gluconobacter suboxydans IFO 3255, membrane-bound D-sorbitol dehydrogenase, that needs product of upstream gene, sldB, for activity. Biosci Biotechnol Biochem 66(11):2314–2322. https://doi.org/10.1271/bbb.66.2314
Siemen A, Kosciow K, Schweiger P, Deppenmeier U (2018) Production of 5-ketofructose from fructose or sucrose using genetically modified Gluconobacter oxydans strains. Appl Microbiol Biotechnol 102(4):1699–1710. https://doi.org/10.1007/s00253-017-8699-1
Singh SK, Banerjee PC (2007) Nucleotide sequence analysis of cryptic plasmid pAM5 from Acidiphilium multivorum. Plasmid 58(2):101–114. https://doi.org/10.1016/j.plasmid.2007.01.005
Soemphol W, Adachi O, Matsushita K, Toyama H (2008) Distinct physiological roles of two membrane-bound dehydrogenases responsible for D-sorbitol oxidation in Gluconobacter frateurii. Biosci Biotechnol Biochem 72(3):842–850. https://doi.org/10.1271/bbb.70720
Soemphol W, Deeraksa A, Matsutani M, Yakushi T, Toyama H, Adachi O, Yamada M, Matsushita K (2011) Global analysis of the genes involved in the thermotolerance mechanism of thermotolerant Acetobacter tropicalis SKU1100. Biosci Biotechnol Biochem 75(10):1921–1928. https://doi.org/10.1271/bbb.110310
Soemphol W, Toyama H, Moonmangmee D, Adachi O, Matsushita K (2007) L-sorbose reductase and its transcriptional regulator involved in L-sorbose utilization of Gluconobacter frateurii. J Bacteriol 189(13):4800–4808. https://doi.org/10.1128/Jb.01895-06
Soisson SM, MacDougall-Shackleton B, Schleif R, Wolberger C (1997) Structural basis for ligand-regulated oligomerization of AraC. Science 276(5311):421–425. https://doi.org/10.1126/science.276.5311.421
Subashini M, Priya AM, Sundarakrishnan B, Jayachandran S (2011) Recombinant Gluconacetobacter diazotrophicus containing cry1Ac gene codes for 130-kDa toxin protein. J Mol Microbiol Biotechnol 20(4):236–242. https://doi.org/10.1159/000331698
Sugiyama M, Suzuki S, Tonouchi N, Yokozeki K (2003) Cloning of the xylitol dehydrogenase gene from Gluconobacter oxydans and improved production of xylitol from D-arabitol. Biosci Biotechnol Biochem 67(3):584–591. https://doi.org/10.1271/bbb.67.584
Sunagawa N, Fujiwara T, Yoda T, Kawano S, Satoh Y, Yao M, Tajima K, Dairi T (2013) Cellulose complementing factor (Ccp) is a new member of the cellulose synthase complex (terminal complex) in Acetobacter xylinum. J Biosci Bioeng 115(6):607–612. https://doi.org/10.1016/j.jbiosc.2012.12.021
Tajima K, Nakajima K, Yamashita H, Shiba T, Munekata M, Takai M (2001) Cloning and sequencing of the beta-glucosidase gene from Acetobacter xylinum ATCC 23769. DNA Res 8(6):263–269. https://doi.org/10.1093/dnares/8.6.263
Takeda Y, Shimizu T (1992) Expression of cytochrome c-553(CO) gene that complements the second subunit deficiency of membrane-bound alcohol dehydrogenase in Gluconobacter suboxydans subsp. α. J Ferment Bioeng 73(2):89–93. https://doi.org/10.1016/0922-338x(92)90319-P
Takeda Y, Shimizu T, Matsushita K, Adachi O, Ameyama M (1992) Role of cytochrome c-553(CO), the second subunit of alcohol dehydrogenase, in the azide-insensitive respiratory chain and in oxidative fermentation of Gluconobacter species. J Ferment Bioeng 74(4):209–213. https://doi.org/10.1016/0922-338x(92)90111-7
Takemura H, Horinouchi S, Beppu T (1993a) Suppression of an ethanol-sensitive mutation of Acetobacter pasteurianus by overexpression of the his1 gene encoding histidinol phosphate aminotransferase. J Ferment Bioeng 76(3):224–228. https://doi.org/10.1016/0922-338x(93)90013-X
Takemura H, Kondo K, Horinouchi S, Beppu T (1993b) Induction by ethanol of alcohol dehydrogenase activity in Acetobacter pasteurianus. J Bacteriol 175(21):6857–6866. https://doi.org/10.1128/jb.175.21.6857-6866.1993
Tang SY, Fazelinia H, Cirino PC (2008) AraC regulatory protein mutants with altered effector specificity. J Am Chem Soc 130(15):5267–5271. https://doi.org/10.1021/ja7109053
Teh MY, Ooi KH, Teo SXD, Bin Mansoor ME, Lim WZS, Tan MH (2019) An expanded synthetic biology toolkit for gene expression control in Acetobacteraceae. ACS Synth Biol 8(4):708–723. https://doi.org/10.1021/j.acssynbio.8b00168
Terpe K (2006) Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol 72(2):211–222. https://doi.org/10.1007/s00253-006-0465-8
Theeragool G, Pitiwittayakul N, Matsutani M, Matsushita K (2018) Disruption of the groEL gene revealed a physiological role for chaperonin in the thermotolerant acetic acid bacterium, Acetobacter pasteurianus SKU1108. Chiang Mai J Sci 45(4):1623–1633
Tonouchi N, Horinouchi S, Tsuchida T, Yoshinaga F (1998a) Increased cellulose production from sucrose by Acetobacter after introducing the sucrose phosphorylase gene. Biosci Biotechnol Biochem 62(9):1778–1780. https://doi.org/10.1271/bbb.62.1778
Tonouchi N, Sugiyama M, Yokozeki K (2003) Construction of a vector plasmid for use in Gluconobacter oxydans. Biosci Biotechnol Biochem 67(1):211–213. https://doi.org/10.1271/bbb.67.211
Tonouchi N, Thara N, Tsuchida T, Yoshinaga F, Beppu T, Horinouchi S (1995) Addition of a small amount of an endoglucanase enhances cellulose production by Acetobacter xylinum. Biosci Biotechnol Biochem 59(5):805–808. https://doi.org/10.1271/bbb.59.805
Tonouchi N, Tsuchida T, Yoshinaga F, Horinouchi S, Beppu T (1994) A host-vector system for a cellulose-producing Acetobacter strain. Biosci Biotechnol Biochem 58(10):1899–1901. https://doi.org/10.1271/bbb.58.1899
Tonouchi N, Yanase H, Kojima Y, Tsuchida T, Yoshinaga F, Horinouchi S (1998b) Increased cellulose production from sucrose with reduced levan accumulation by an Acetobacter strain harboring a recombinant plasmid. Biosci Biotechnol Biochem 62(5):833–836. https://doi.org/10.1271/bbb.62.833
Trcek J, Mira NP, Jarboe LR (2015) Adaptation and tolerance of bacteria against acetic acid. Appl Microbiol Biotechnol 99(15):6215–6229. https://doi.org/10.1007/s00253-015-6762-3
Urakami T, Tamaoka J, Suzuki K, Komagata K (1989) Acidomonas gen. nov., incorporating Acetobacter methanolicus as Acidomonas methanolica comb. nov. Int J Syst Bacteriol 39(1):50–55. https://doi.org/10.1099/00207713-39-1-50
Urzua LS, Vazquez-Candanedo AP, Sanchez-Espindola A, Ramirez CA, Baca BE (2013) Identification and characterization of an iron ABC transporter operon in Gluconacetobacter diazotrophicus PAL5. Arch Microbiol 195(6):431–438. https://doi.org/10.1007/s00203-013-0890-x
Valero F (2012) Heterologous expression systems for lipases: A review. Methods Mol Biol 861:161–178. https://doi.org/10.1007/978-1-61779-600-5_11
Valla S, Coucheron DH, Fjaervik E, Kjosbakken J, Weinhouse H, Ross P, Amikam D, Benziman M (1989) Cloning of a gene involved in cellulose biosynthesis in Acetobacter xylinum - Complementation of cellulose-negative mutants by the UDPG pyrophosphorylase structural gene. Mol Gen Genet 217(1):26–30. https://doi.org/10.1007/Bf00330938
Valla S, Coucheron DH, Kjosbakken J (1986) Conjugative transfer of the naturally-occurring plasmids of Acetobacter xylinum by IncP-plasmid-mediated mobilization. J Bacteriol 165(1):336–339. https://doi.org/10.1128/jb.165.1.336-339.1986
Valla S, Coucheron DH, Kjosbakken J (1987) The plasmids of Acetobacter xylinum and their interaction with the host chromosome. Mol Gen Genet 208(1-2):76–83. https://doi.org/10.1007/Bf00330425
Varotto-Boccazzi I, Epis S, Arnoldi I, Corbett Y, Gabrieli P, Paroni M, Nodari R, Basilico N, Sacchi L, Gramiccia M, Gradoni L, Tranquillo V, Bandi C (2020) Boosting immunity to treat parasitic infections: Asaia bacteria expressing a protein from Wolbachia determine M1 macrophage activation and killing of Leishmania protozoans. Pharmacol Res 105288. https://doi.org/10.1016/j.phrs.2020.105288
Velazquez-Hernandez ML, Baizabal-Aguirre VM, Cruz-Vazquez F, Trejo-Contreras MJ, Fuentes-Ramirez LE, Bravo-Patino A, Cajero-Juarez M, Chavez-Moctezuma MP, Valdez-Alarcon JJ (2011) Gluconacetobacter diazotrophicus levansucrase is involved in tolerance to NaCl, sucrose and desiccation, and in biofilm formation. Arch Microbiol 193(2):137–149. https://doi.org/10.1007/s00203-010-0651-z
Voss J, Ehrenreich A, Liebl W (2010) Characterization and inactivation of the membrane-bound polyol dehydrogenase in Gluconobacter oxydans DSM 7145 reveals a role in meso-erythritol oxidation. Microbiol 156:1890–1899. https://doi.org/10.1099/mic.0.037598-0
Walker KT, Goosens VJ, Das A, Graham AE, Ellis T (2019) Engineered cell-to-cell signalling within growing bacterial cellulose pellicles. Microbiol Biotechnol 12(4):611–619. https://doi.org/10.1111/1751-7915.13340
Wang B, Shao YC, Chen FS (2015) Overview on mechanisms of acetic acid resistance in acetic acid bacteria. World J Microb Biotechnol 31(2):255–263. https://doi.org/10.1007/s11274-015-1799-0
Wang PP, Xia Y, Li JH, Kang Z, Zhou JW, Chen J (2016) Overexpression of pyrroloquinoline quinone biosynthetic genes affects L-sorbose production in Gluconobacter oxydans WSH-003. Biochem Eng J 112:70–77. https://doi.org/10.1016/j.bej.2016.04.011
Wei LJ, Yang XP, Gao KL, Lin JP, Yang SL, Hua QA, Wei DZ (2010) Characterization of enzymes in the oxidation of 1,2-propanediol to D-(-)-lactic acid by Gluconobacter oxydans DSM 2003. Mol Biotechnol 46(1):26–33. https://doi.org/10.1007/s12033-010-9263-8
Wei LJ, Zhou JL, Zhu DN, Cai BY, Lin JP, Hua Q, Wei DZ (2012) Functions of membrane-bound alcohol dehydrogenase and aldehyde dehydrogenase in the bio-oxidation of alcohols in Gluconobacter oxydans DSM 2003. Biotechnol Bioproc Eng 17(6):1156–1164. https://doi.org/10.1007/s12257-012-0339-0
Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, Benziman M, Gelfand DH, Meade JH, Emerick AW, Bruner R, Benbassat A, Tal R (1990) Genetic organization of the cellulose synthase operon in Acetobacter xylinum. P Natl Acad Sci USA 87(20):8130–8134. https://doi.org/10.1073/pnas.87.20.8130
Wu XF, Yao HL, Cao LL, Zheng Z, Chen XJ, Zhang M, Wei ZJ, Cheng JS, Jiang ST, Pan LJ, Li XJ (2017) Improving acetic acid production by over-expressing PQQ-ADH in Acetobacter pasteurianus. Front Microbiol 8. ARTN 1713. https://doi.org/10.3389/fmicb.2017.01713
Xu S, Wang XB, Du GC, Zhou JW, Chen J (2014) Enhanced production of L-sorbose from D-sorbitol by improving the mRNA abundance of sorbitol dehydrogenase in Gluconobacter oxydans WSH-003. Microbiol Cell Fact 13. ARTN 146. https://doi.org/10.1186/s12934-014-0146-8
Yadav V, Paniliatis BJ, Shi H, Lee K, Cebe P, Kaplan DL (2010) Novel in vivo-degradable cellulose-chitin copolymer from metabolically engineered Gluconacetobacter xylinus. Appl Environ Microbiol 76(18):6257–6265. https://doi.org/10.1128/Aem.00698-10
Yakushi T, Fukunari S, Kodama T, Matsutani M, Nina S, Kataoka N, Theeragool G, Matsushita K (2018a) Role of a membrane-bound aldehyde dehydrogenase complex AldFGH in acetic acid fermentation with Acetobacter pasteurianus SKU1108. Appl Microbiol Biotechnol 102(10):4549–4561. https://doi.org/10.1007/s00253-018-8940-6
Yakushi T, Komatsu K, Matsutani M, Kataoka N, Vangnai AS, Toyama H, Adachi O, Matsushita K (2018b) Improved heterologous expression of the membrane-bound quinoprotein quinate dehydrogenase from Gluconobacter oxydans. Protein Expres Purif 145:100–107. https://doi.org/10.1016/j.pep.2018.01.007
Yakushi T, Takahashi R, Matsutani M, Kataoka N, Hours RA, Ano Y, Adachi O, Matsushita K (2020) The membrane-bound sorbosone dehydrogenase of Gluconacetobacter liquefaciens is a pyrroloquinoline quinone-dependent enzyme. Enzyme Microbiol Tech 137. ARTN 109511. https://doi.org/10.1016/j.enzmictec.2020.109511
Yakushi T, Terada Y, Ozaki S, Kataoka N, Akakabe Y, Adachi O, Matsutani M, Matsushita K (2018c) Aldopentoses as new substrates for the membrane-bound, pyrroloquinoline quinone-dependent glycerol (polyol) dehydrogenase of Gluconobacter sp. Appl Microbiol Biotechnol 102(7):3159–3171. https://doi.org/10.1007/s00253-018-8848-1
Yamada Y (2014) Transfer of Gluconacetobacter kakiaceti, Gluconacetobacter medellinensis and Gluconacetobacter maltaceti to the genus Komagataeibacter as Komagataeibacter kakiaceti comb. nov., Komagataeibacter medellinensis comb. nov. and Komagataeibacter maltaceti comb. nov. Int J Syst Evol Microbiol 64(Pt 5):1670–1672. https://doi.org/10.1099/ijs.0.054494-0
Yamada Y, Hoshino K, Ishikawa T (1997) The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA: the elevation of the subgenus Gluconoacetobacter to the generic level. Biosci Biotechnol Biochem 61(8):1244–1251. https://doi.org/10.1271/bbb.61.1244
Yamada Y, Yukphan P, Lan Vu HT, Muramatsu Y, Ochaikul D, Tanasupawat S, Nakagawa Y (2012) Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J Gen Appl Microbiol 58(5):397–404. https://doi.org/10.2323/jgam.58.397
Yang XP, Wei LJ, Lin JP, Yin B, Wei DZ (2008) Membrane-bound pyrroloquinoline quinone-dependent dehydrogenase in Gluconobacter oxydans M5, responsible for production of 6-(2-hydroxyethyl) amino-6-deoxy-L-sorbose. Appl Environ Microbiol 74(16):5250–5253. https://doi.org/10.1128/Aem.00272-08
Yuan JF, Wu MB, Lin JP, Yang LR (2016a) Combinatorial metabolic engineering of industrial Gluconobacter oxydans DSM2343 for boosting 5-keto-D-gluconic acid accumulation. BMC Biotechnol 16. ARTN 42. https://doi.org/10.1186/s12896-016-0272-y
Yuan JF, Wu MB, Lin JP, Yang LR (2016b) Enhancement of 5-keto-D-gluconate production by a recombinant Gluconobacter oxydans using a dissolved oxygen control strategy. J Biosci Bioeng 122(1):10–16. https://doi.org/10.1016/j.jbiosc.2015.12.006
Zahid N, Deppenmeier U (2016) Role of mannitol dehydrogenases in osmoprotection of Gluconobacter oxydans. Appl Microbiol Biotechnol 100(23):9967–9978. https://doi.org/10.1007/s00253-016-7680-8
Zeng WZ, Cai W, Liu L, Du GC, Chen J, Zhou JW (2019) Efficient biosynthesis of 2-keto-D-gluconic acid by fed-batch culture of metabolically engineered Gluconobacter japonicus. Synth Syst Biotechnol 4(3):134–141. https://doi.org/10.1016/j.synbio.2019.07.001
Zhang H, Shi LL, Lin JP, Sun M, Wei DZ (2016) Effective improvement of the activity of membrane-bound alcohol dehydrogenase by overexpression of adhS in Gluconobacter oxydans. Biotechnol Lett 38(7):1131–1138. https://doi.org/10.1007/s10529-016-2084-5
Zhang JL, Li S, Xu H, Zhou P, Zhang LJ, Ouyang PK (2013) Purification of xylitol dehydrogenase and improved production of xylitol by increasing XDH activity and NADH supply in Gluconobacter oxydans. J Agr Food Chem 61(11):2861–2867. https://doi.org/10.1021/jf304983d
Zhang L, Lin JP, Ma YS, Wei DZ, Sun M (2010) Construction of a novel shuttle vector for use in Gluconobacter oxydans. Mol Biotechnol 46(3):227–233. https://doi.org/10.1007/s12033-010-9293-2
Zhang XL, Zheng Y, Xia ML, Wu YN, Liu XJ, Xie SK, Wu YF, Wang M (2020) Knowledge domain and emerging trends in vinegar research: A bibliometric review of the literature from WoSCC. Foods 9(2) ARTN 166. https://doi.org/10.3390/foods9020166
Zheng Y, Wang J, Bai XL, Chang YG, Mou J, Song J, Wang M (2018) Improving the acetic acid tolerance and fermentation of Acetobacter pasteurianus by nucleotide excision repair protein UvrA. Appl Microbiol Biotechnol 102(15):6493–6502. https://doi.org/10.1007/s00253-018-9066-6
Zhu HX, Jia SR, Yang HJ, Yan L, Li J (2012) The study of optimal conditions of electroporation in Gluconacetobacter xylinum. Adv Intel Soft Compu 115:17–24
Zhu JW, Xie JL, Wei LJ, Lin JP, Zhao L, Wei DZ (2018) Identification of the enzymes responsible for 3-hydroxypropionic acid formation and their use in improving 3-hydroxypropionic acid production in Gluconobacter oxydans DSM 2003. Bioresource Technol 265:328–333. https://doi.org/10.1016/j.biortech.2018.06.001
Zhu K, Lu LF, Wei LJ, Wei DZ, Imanaka T, Hua Q (2011) Modification and evolution of Gluconobacter oxydans for enhanced growth and biotransformation capabilities at low glucose concentration. Mol Biotechnol 49(1):56–64. https://doi.org/10.1007/s12033-011-9378-6
Zou XX, Lin JP, Mao XL, Zhao SY, Ren YH (2017) Biosynthesis of L-erythrose by assembly of two key enzymes in Gluconobacter oxydans. J Agr Food Chem 65(35):7721–7725. https://doi.org/10.1021/acs.jafc.7b02201
Acknowledgements
The authors would like to thank the central library of the Forschungszentrum Jülich for their support.
Funding
Open Access funding enabled and organized by Projekt DEAL. We are grateful to the Federal Ministry of Education and Research (BMBF) for the financial support of PF within the project IMPRES (031B0370B). The funding organization did not influence the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
PMF, AK, and TP screened and selected the literature. All authors developed the manuscript, revised the manuscript, and read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fricke, P.M., Klemm, A., Bott, M. et al. On the way toward regulatable expression systems in acetic acid bacteria: target gene expression and use cases. Appl Microbiol Biotechnol 105, 3423–3456 (2021). https://doi.org/10.1007/s00253-021-11269-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11269-z