Abstract
Citric acid production by Aspergillus niger and itaconic acid production by Aspergillus terreus are two major examples of technical scale fungal fermentations based on metabolic overflow of primary metabolism. Both organic acids are formed by the same metabolic pathway, but whereas citric acid is the end product in A. niger, A. terreus performs two additional enzymatic steps leading to itaconic acid. Despite of this high similarity, the optimization of the production process and the mechanism and regulation of overflow of these two acids has mostly been investigated independently, thereby ignoring respective knowledge from the other. In this review, we will highlight where the similarities and the real differences of these two processes occur, which involves various aspects of medium composition, metabolic regulation and compartmentation, transcriptional regulation, and gene evolution. These comparative data may facilitate further investigations of citric acid and itaconic acid accumulation and may contribute to improvements in their industrial production.
Similar content being viewed by others
Introduction
Citric acid (2-hydroxy-propane-1,2,3-tricarboxylic acid) and itaconic acid (2-methylene-succinic acid or 2-methylidenebutanedioic acid) are the most well-known examples of fungal “overflow metabolism,” a term coined by Foster (1949) to describe the seemingly wasteful strategy of some fungi to incompletely oxidize their carbon source. The first fungal production process for citric acid employing Aspergillus niger is a century old (Currie 1917). Itaconic acid accumulation by Aspergillus itaconicus was described in the early 30s of the last century (Kinoshita 1931), while the first production technology—already employing Aspergillus terreus—was patented in the next decade (Kane et al. 1945). Before World War II, organic acid manufacturing was exclusively performed by the labor-intensive and relatively low-yield surface method (Doelger and Prescott 1934; Calam et al. 1939). In this process, fungi were grown in aluminum trays of 50–200 cm depth, filled up with sugar and inorganic nutrients, with sterile air—serving as an oxygen supply and a cooling agent—being passed over them. Submerged organic acid fermentation employing agitated, aerated, high-quality steel vessels started on the sidelines of penicillin manufacturing (Kane et al. 1945; Perlman et al. 1946a, b; Lockwood and Nelson 1946; Martin and Waters 1952). Commercial large-scale submerged fermentations by A. niger (for citric acid) and A. terreus (for itaconic acid) have been initiated all across the world in the 50s (Steel et al. 1955). Although some citric acid is produced by solid-state fermentation, the estimated 2.1 million tons of citric acid manufactured annually is almost completely performed by the submerged method, most typically batch wise (Cavallo et al. 2017). Itaconic acid is exclusively produced this way (Bafana and Pandey 2018). Despite the intense research on lab- and pilot scale (Yu et al. 2018), no continuous or immobilized cell-based fermentation methods are used on technical scale for any of the two organic acids. In summary, from the technological point of view (scale, vessel types, inoculation protocols, critical process parameters, medium composition), the upstream parts as well as kinetics of product and biomass formation of these two industrial fermentations are very much similar (Supplementary Tables S1 and S2).
Citric acid is mainly used as a flavoring agent in the food industry, and to a lesser extent as an acidifier and a chelating agent in the chemical and pharma industries (Apelblat 2014). Itaconic acid is produced in significantly less amounts (41.000 t/a in 2011; Geiser et al. 2016), but the actual market potential is estimated at 80.000 t/a. Indeed, in 2004, the US Department of Energy assigned itaconic acid as one of the top 12 most promising building block chemicals for bio-based economy (Werpy and Petersen 2004). Due to the conjugated double bond of the methylene group, its fundamental application is being a precursor for the polymer industry (absorbents, unsaturated polyester resins, plastics; Okabe et al. 2009; Kuenz and Krull 2018). Importantly, itaconic acid could in principle replace polyacrylic acid whose production is petroleum-based. Should that happen completely, a market worth over $11 billion would open up (El-Imam and Du 2014; Saha et al. 2018), making itaconic acid one of the crown jewels of industrial biotechnology. However, that would require at least 25% fall in prices that currently stands at around $2/kg, preventing wider use of this compound.
The biochemistry and production of citric and itaconic acid have recently been subject of several excellent reviews (Steiger et al. 2013; Chen and Nielsen 2016; Bafana and Pandey 2018; Zhao et al. 2018; Kuenz and Krull 2018), but a critical comparison of the similar and dissimilar aspects of their biosynthesis and accumulation has not been made. In this review, we will document that the biochemistry and physiology of accumulation of these two organic acids is indeed almost identical. The only difference is the presence of three additional genes in A. terreus, whose evolution and origin will be discussed.
Biological importance of citric acid and itaconic acid
The biological function of the formation of the two acids have never been studied in details, but it is generally assumed that both serve as strong acidifiers and therefore provide selective advance for these two acid-tolerant fungi in their habitat over bacteria and yeast. Another potential function of citric acid (but not itaconic acid) may be its ability to chelate manganese and ferrous ions, thereby participating in their transport into the cell and conversely, in the scavenging of these metal ions from uptake by competing organisms (Guerinot et al. 1990; Odoni et al. 2017). Out-competing rivals for metal ions has been shown to be an important defense strategy in fungi (Canessa and Larrondo 2013; Blatzer and Latgé 2017).
In contrast to citric acid, itaconic acid itself has been described as a weapon to combat competitors: itaconic acid has been identified in activated pro-inflammatory macrophages, and its effect on cellular metabolism has been attributed to the inhibition of succinate dehydrogenase (Strelko et al. 2011; Michelucci et al. 2013). Furthermore, itaconate was shown to induce electrophilic stress through reacting with glutathione and subsequently inducing the transcription factor Nrf2 which is essential for protection against oxidative and xenobiotic stresses, and attenuate inflammation (Kobayashi et al. 2013; Bambouskova et al. 2018). Itaconate also inhibits isocitrate lyase (McFadden et al. 1971), a key enzyme of the glyoxylate cycle, required for the survival of bacterial and fungal pathogens inside the eukaryotic host (Sasikaran et al. 2014). Although the role of itaconic acid in the survival of A. terreus in its natural environment has not been studied yet, we consider reasonable that its antimicrobial effects are beneficial also there, and that this could have an impact on the regulation of its formation.
Biochemical pathways of citric and itaconic acid biosynthesis
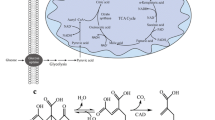
Citric acid is an intermediate of the citric acid cycle, which is an essential part of energy metabolism in all aerobic eukaryotic heterotrophs (Krebs 1940). Cleland and Johnson (1954), using 14C-labeled d-glucose pioneered by demonstrating that citric acid accumulation occurs by a condensation of oxaloacetate and pyruvate, the former arising by fixation of the carbon dioxide from one of the two pyruvate molecules derived from glucose during oxidative decarboxylation to acetyl-CoA. By this way, no carbon dioxide is lost what enabled molar yields of citric acid > 66%, the maximum that would arise by the standard operation of the TCA cycle. They also demonstrated that no further metabolism of citric acid by the citric acid cycle occurred, and that no CO2 was released by the pentose phosphate pathway.
Shortly thereafter, Bentley and Thiessen (1957) demonstrated that the same pathway operates in A. terreus with the only exception that citric acid is further metabolized via cis-aconitate to itaconate. The operation of this pathway in an industrial strain of A. terreus was later confirmed by tracer experiments with 14C- and 13C-labeled substrates (Bonnarme et al. 1995). They also showed that no CO2 was released during the production phase, indicating that no glucose metabolism via the pentose phosphate pathway occurred during itaconic acid accumulation either.
A further point that must be noted is that citrate is a product of incomplete oxidative metabolism, and therefore contains one more oxygen atom than the hexose from which it is formed (Fig. 1). This also applies to itaconic acid, although the overall stoichiometry discloses this because of the removal of carbon dioxide toward the end of its biosynthesis. This requirement of extra oxygen has important implications for the conditions leading to the production of these two acids in high amounts (see below). To sum up, the only metabolic difference between citric acid and itaconic acid biosynthesis is that A. terreus does not accumulate citrate but metabolizes it to cis-aconitate, which is then converted to itaconate by cis-aconitate decarboxylase (Steiger et al. 2013).
Nutrient preconditions for citric acid and itaconic acid production
To discuss similarities and dissimilarities in the production of citric acid and itaconic acid, the first aspect that needs to be compared is the nutrient requirements for these two processes. The fundamental prerequisite for overflow metabolism is a metabolic deregulation that is caused by a combination of abundance of the carbon source and serious shortage of phosphate and some micronutrient(s). Yet there are other, critical factors that determine whether the accumulation of citric and itaconic acid reaches levels beyond a molar Yp/s > 0.7 (i.e., by a process involving pyruvate carboxylation, see above). Studies that fulfilled this criterion showed that the carbon source must be a rapidly metabolizable hexose or disaccharide and be supplied in concentrations > 100 g/L. This high concentration of the hexose obviously also requires a correspondingly high degree of aeration to fulfill the stoichiometry of citric acid/itaconic acid biosynthesis (see above; Molnár et al. 2018; Figs. 2 and 3). The concentration of inorganic nutrients should be sufficient to enable biomass formation in amounts of 10–15 g dry weight/L, which enables the production of both acids in the range of yields defined above.
A 3D plot demonstrating the trilateral relationship between citric acid molar yield, aeration of the fermenter, and the concentration of the carbon source used. Input data are taken from well-documented, publicly available fermentations, listed in Supplementary Table S5. Plot was made with SigmaPlot 12.0 (Systat Software, Inc., USA)
A 3D plot demonstrating the trilateral relationship between itaconic acid molar yield, aeration of the fermenter, and the concentration of the carbon source used. Input data are taken from well-documented, publicly available fermentations, listed in Supplementary Table S6. Plot was made with the same software as above
In addition, both acids have been proven to be accumulated in the respective high concentrations only if the pH of the medium is below 3. There are some data in the literature that itaconic acid can also be produced at a higher pH (Nubel and Ratajak 1962; Hevekerl et al. 2014), but these studies did not reach the high itaconic acid concentrations we consider essential for this review. In A. niger, the requirement for a low pH has been explained by the need to bypass gluconic acid formation by the extracellular glucose oxidase which is inactivated by a pH < 3 (Kubicek and Karaffa 2010), but A. terreus is not known to produce gluconic acid. The reason for the requirement of a low pH must therefore be due to different, so far unknown reasons.
Conflicting information is available about the role of metal ions for citric and itaconic acid formation, respectively. In the case of A. niger, US and Canadian research groups led by Johnson and Clark, respectively, have demonstrated that high yields can only be obtained when the concentration of Mn(II) ions in the medium is below 4 μg/L (Clark 1962; Fig. 4). A similar inhibitory effect has been demonstrated for Fe(II) at concentrations > 2 mg/L (Tomlinson et al. 1950). The concentration of Zn(II) did not affect citric acid formation. Interestingly, the inhibition of citric acid accumulation by iron has attracted more attention in literature. In our opinion, this has two main reasons: one is that purification of molasses by the addition of hexacyanoferrate—a classical method to remove ferrous ions—has been proven in industry to be necessary to achieve high citric acid yields. The other one is that the first enzyme that performs downstream metabolism of citrate in the cell—aconitase—is an iron-containing enzyme, and it was believed for a long time that iron limitation in the medium leads to a blockage of aconitase activity and thus triggers citric acid overflow (Ramakrishnan et al. 1955). Yet this hypothesis has been clearly falsified by proving that citrate and isocitrate are present in citric acid-producing mycelia of A. niger in concentrations perfectly reflecting the aconitase equilibrium and the enzyme must therefore be active (Kubicek and Röhr 1985).
A 3D plot demonstrating the trilateral relationship between citric acid molar yield, Mn(II) ion concentration, as well as carbon source concentration used. Input data are taken from well-documented, publicly available fermentations, listed in Supplementary Table S7. Plot was made with the same software as above
How can the effect of iron then be interpreted? The critical point is that metal ions of analytical grade, as used in research, always contain impurities. Iron salts contain up to 0.5% (w/w) manganese, which implies that the addition of 2 mg/L iron introduces a concentration of 10 μg/L of Mn(II) ions, which—as explained above—would per se be sufficient for a significant inhibition of citric acid formation. In addition, the treatment of molasses with ferrocyanide precipitates not only iron but also manganese and zinc ions. This suggests that the effects reported for iron are in fact due to the presence of manganese ions, which therefore is the critical parameter to obtain high yield of citrate.
With regard to itaconic acid accumulation, comparably few studies in this direction have been published. Relevant literature regarding the effect of Mn(II) ions is summarized in Fig. 5. Ferrocyanide treatment was shown to enhance itaconic acid production (Batti and Schweiger 1963). Karaffa et al. (2015) provided evidence that the overproduction of itaconic acid requires a medium in which the Mn(II) concentration is in the same range as in citric acid fermentations. Interestingly, however, there are also reports on itaconic acid fermentations with high yields which do not attempt to remove metal ions (Shin et al. 2013, 2017). This issue therefore needs further investigation.
A 3D plot demonstrating the trilateral relationship between itaconic acid molar yield, Mn(II) ion concentration, as well as carbon source concentration used. Input data are taken from well-documented, publicly available fermentations, listed in Supplementary Table S8. Plot was made with the same software as above
Morphology
The above described manganese deficiency also leads to a typical restricted morphology which consists of small pellets (< 0.5 mm diameter) with branches of short hyphae with swollen tips (Snell and Schweiger 1949; Takahashi et al. 1958; Takahashi and Yamada 1959). Image analysis showed that citric acid molar yields exceed 85% when the macro-morphological form is constituted by stable particles of intertwined filaments around a small nucleus, while the micro-morphology is characterized by branches of short hyphae with swollen tips (Cox et al. 1998; Papagianni and Mattey 2006). A similar morphology was developed by A. terreus during high-yield itaconate production (Batti and Schweiger 1963; Gyamerah 1995; Kuenz et al. 2012). Karaffa et al. (2015) showed that Mn(II) deficiency is the critical factor determining this morphology also in A. terreus. Thus, the influence of manganese and other heavy metal ions on the morphology is similar in citric acid and itaconic acid fermentations. The background of this requirement is believed to be the decreased viscosity of the fermentation broth, which results in improved oxygen transfer and increased solute uptake.
There have been few attempts only to genetically stabilize the development of the small pellets, and they have so far been done with A. niger only: the main cell wall of A. niger contains β-1,3-glucan, chitin, β-1,6-glucan, α-1,3-glucan, galactosaminogalactan, and galactomannan (Pel et al. 2007). The cell walls of the pellets that are formed under manganese-deficient conditions have an increased chitin and lowered ß-glucan content (Kisser et al. 1980). Consistent with this, Yin et al. (2017) found that expression of chitin synthase C (chsC) and pstA, encoding a cell surface glycosylphosphatidylinositol-anchored protein important for cell wall integrity and resistance against low pH (Pardo et al. 2004; Gil-Bona et al. 2015), were highly upregulated during citric acid fermentation. Sun et al. (2018) silenced chsC in an industrial mutant strain which resulted in strains with altered morphology and with lower proportion of dispersed mycelia, which caused the desired decrease in viscosity, improved oxygen, and mass transfer efficiency and improved citric acid production. Dai et al. (2004) applied a different strategy (suppression subtractive hybridization) to identify genes that are differentially expressed under manganese sufficient and deficient conditions, and identified a gene in A. niger (brsa25 = An03g00640), which encodes an unknown amino acid permease whose antisense inhibition permitted pelleted growth and increased citrate production also in the presence of high manganese concentrations.
Evolution of the itaconate biosynthesis cluster
The identification of the genes encoding the necessary steps leading from citrate to itaconate in the A. terreus genome was a breakthrough in the understanding of itaconic acid biosynthesis. Using differential expression, Li et al. (2011a, b) identified four genes—one (abbreviation still lacking) encoding a Zn2Cys6-type transcriptional regulator, mttA (encoding a mitochondrial cis-aconitate transporter), cadA (cis-aconitate decarboxylase), and mfsA encoding a membrane transporter of the major facilitator subfamily—that occur adjacent to the cluster for the biosynthesis of the polyketide lovastatin (Fig. 6). Two further genes—cypA (encoding a cytochrome p450 monooxygenase) and rdoA (encoding an α-ketoglutarate-(FeII)-dependent dioxygenase)—are located at the 3′ side of the itaconic acid cluster. They could be involved in the conversion of itaconic acid to 2-hydroxiparaconic acid which involves further oxidation steps (Guevarra and Tabuchi 1990). It is tempting to speculate that itaconic acid formation originated from this cluster, e.g., by decreasing the expression of cypA and rdoA. Unfortunately, the expression of these two genes has not been studied under itaconic acid-producing conditions yet. In this review, we therefore use the term “itaconate gene cluster” only for the four genes first identified by Li et al. (2011a, b). CadA and mttA were among the most strongly expressed under the conditions used by the authors. Interestingly, functional verification of these four genes by gene deletion is still lacking, and experimental evidence for the role of cadA, mttA, and mfsA in itaconic acid biosynthesis has been provided only by introducing them into A. niger which lacks cadA and mttA and therefore does not produce itaconic acid. Consequently, the introduction of cadA alone lead only to very low titers of itaconic acid formation, whereas the introduction of cadA and mttA led to the much higher titers (Li et al. 2011a, b; van der Straat et al. 2014; van der Straat and de Graaff 2014), indicating that these are the critical elements discriminating the citric acid and itaconic acid overflow. Introduction of mfsA (either with cadA or together with cadA and mttA) did not lead to increased production levels of itaconic acid, implying that an endogenous A. niger transporter can efficiently transport itaconate. Interestingly, expression of mfsA in A. niger led to increased citrate production, suggesting that MfsA may be a transporter with broad substrate specificity. Broad substrate specificity was demonstrated for many transporters of the MFS superfamily (Yan 2013) and MfsA orthologs are present in several other Aspergilli who do not produce itaconic acid (Supplementary Table S3). It would therefore be interesting to investigate the phenotype of an mfsA knock-out strain of A. terreus with respect to itaconic acid production.
Genomic environment of the Zn2Cys6 transcriptional activator present in the itaconate biosynthesis cluster of A. terreus and in A. niger. Genes are specified by the protein ID used in A. terreus NIH 2624 (https://genome.jgi.doe.gov/Aspte1/Aspte1.home.html) and A. niger ATCC 1015 (https://genome.jgi.doe.gov/Aspni7/Aspni7.home.html). Italic abbreviations specify the genes whose function has been experimentally verified
The two essential enzymes for itaconate production—CadA and MttA—seem to be absent from other fungi, because the best hits in NCBI BLASTp are proteins with only low score, low overall similarity, and a high E value (Supplementary Table S3). It is therefore interesting that the relatively best hits for MttA were not found in other Aspergilli, but are scattered throughout different other fungal classes and families. This could either indicate that MttA arose by horizontal gene transfer or that an ancient progenor of the mttA gene was lost in all other Aspergilli. In contrast, the best hits for CadA—although of low score, identity, and high E values—were exclusively from Aspergilli and Penicillia. Interestingly, most of them were annotated as methylcitrate dehydratases, which are members of the same protein family (MmgE/PrpD) as CadA. The close similarity of these dehydratases with carboxylase is puzzling: the decarboxylation reaction catalyzed by CadA is unique among carboxy-lyases because it catalyzes a decarboxylation resulting in both the reduction of a β-γ unsaturation as well as the formation of a methylidene group α by decarboxylation without removal of any other group (Strelko et al. 2011). A phylogeny of the best hits of CadA in NCBI BLASTp shows that it forms a basal clade to all other putative methylcitrate dehydratases with three undefined Penicillium proteins as a sister clade (Fig. 7). CadA therefore seems to have evolved before the splitting of Aspergillus and Penicillium (200–150 mya; van Steenwyk et al. 2018) and subsequently has been lost in most other species of these two fungal families. The catalytic nature of the progenor of the MmgE/PrpD family remains obscure, however. A cis-aconitate decarboxylase has also been identified in mammalian cells (where it is called immune-responsive gene 1, Irg1), but the IRG1 protein also catabolizes fatty acid-derived metabolites (Michelucci et al. 2013). Despite catalyzing cis-aconitate decarboxylation, no homology could be detected between IRG1 and CadA (L. Karaffa and C.P. Kubicek, unpublished data).
Maximum likelihood tree of A. terreus CadA (cis-aconitate decarboxylase). Proteins used for the tree were identified by a BLASTp search at NCBI, using CadA as bait and the 32 closest hits used for a phylogenetic analysis using maximum likelihood with 1000 bootstrap replicase in MEGA5.0 (https://www.megasoftware.net/). With the exception of CadA, none of the detected proteins has been functionally characterized. The names used in the tree are those submitted by the depositors and should be considered hypothetical
Interestingly, the gene encoding the Zn2Cys6 transcriptional activator of the A. terreus “itaconate cluster” is present in other Aspergilli including A. niger (E value 2E-119; 42% identity). In the A. niger genome, it clusters with genes encoding a cytosolic citrate synthase (citB), a methylcitrate dehydratase, a fatty acid synthase, and a putative terpenoid synthase (Fig. 6). Hossain et al. (2016) proposed that this gene cluster may encode the genes for the biosynthesis of tensyuic acid, an alkyl-itaconic acid (Hasegawa et al. 2007). This would imply that A. niger can form itaconate but obviously by a different mechanism: the gene encoding methylcitrate synthase in this cluster (XP_001393190.1) shares no major similarity with CadA and its close hits in BLASTp all annotate as methylcitrate dehydratase for which activity on cis-aconitate has so far not been reported yet. The role of the Zn2Cys6 transcription factor present in the “itaconic acid” and “tensyuic acid cluster,” with respect to itaconic and citric acid accumulation, clearly needs further studies. Transformation of Aspergillus nidulans, who does not produce citric acid, with A. niger citB has recently been shown to lead to citric acid accumulation (Vesth et al. 2018) thus stressing the potential importance of this gene in this process.
Finally, we should like to note that the genes necessary for the formation of itaconic acid by Ustilago mayidis are also organized as a cluster in the genome (Geiser et al. 2016). However, its biosynthesis involves trans- not cis-aconitate, and BLASTp of the A. terreus genome with the U. mayidis cluster genes identifies only genes not occurring in the “itaconate cluster” as best hits (although with less than 41% identity and E values > E-040). Since the clusters in these two fungi do not comprise homologs, they are likely the result of convergent evolution.
Glycolytic flux and the role of phosphofructokinase
The tracer studies emphasized above show that the almost exclusive operation of glycolysis plays a major role in both processes, and that its speed could be rate limiting. Based on studies with Saccharomyces cerevisiae, metabolic regulation by allosteric feedback inhibition and feed-forward activation was accepted as a major mechanism controlling the flux through glycolysis in fungi, and 6-phosphofructokinase (PfkA) was considered as the candidate for the strongest control (Fiechter and Gmünder 1989; Entian and Barnett 1992). However, regulation at this step during citric acid production was challenged by the findings that citrate is a metabolic inhibitor of PfkA from yeast (Salas et al. 1965).
The kinetic properties of PfkA have therefore been extensively studied in A. niger. It is upregulated by fructose-2,6-biphosphate (Fru-2,6-P2), AMP, and NH4+ ions, while ATP and citrate have inhibitory effects on the enzyme (Habison et al. 1983; Arts et al. 1987). Since the intracellular Fru-2,6-P2 concentration increased considerably with increasing concentrations of glucose in the medium (Kubicek et al. 1990), Fru-2,6-P2 may antagonize citrate inhibition in vivo. This hypothesis is consistent with the findings that the overexpression of pfkA gene in A. niger did not increase citric acid production, but reduced the intracellular concentration of Fru-2,6-P2 almost twofold compared to the wild-type strain. Thus, the fungus seems to adapt to overexpression of phosphofructokinase by decreasing the specific activity of the enzyme through a reduction in the level of fructose 2,6-P2.
In addition, it was observed (Mlakar and Legiša 2006; Capuder et al. 2009) that PfkA undergoes proteolytic truncation when A. niger is grown under citric acid-producing conditions, and that this fragment (after phosphorylation by protein kinase A) is not inhibited by citrate. Interestingly, this PfkA fragment is stimulated by Fru-2,6-P2 to a higher degree than the native PfkA, thus enabling a higher activity in vivo. Consistent with these interpretations, transformation of A. niger with this PFK fragment resulted in higher rates of citric acid production (Capuder et al. 2009).
The successful stimulation of citric acid production in A. niger by a PfkA fragment prompted to test whether this fragment would also increase the rate of itaconic acid production by A. terreus. Its expression in A. terreus (Tevz et al. 2010) resulted in an increased rate of itaconate accumulation and a 1.5-fold increase in its final concentration. Unfortunately, however, the parent strain formed only 28 g/L itaconate (Yp/s = 0.4). Huang et al. (2014), performing similar experiments, also observed a twofold stimulation of itaconic acid production but only under conditions of low final molar yield (Yp/s = 0.15 in the parent strain), whereas no effect was observed in a strain displaying high molar yields (Yp/s = 0.9). This difference could be explained by the fact that the latter study used an industrial strain of A. terreus in which glycolytic flux may already have been enhanced during strain development.
One point not considered so far is why a citrate-insensitive PfkA would lead to increased glycolytic rates during itaconate production at all. During citric acid production by A. niger, citrate is transported into the cytoplasm where it reaches high levels (Habison et al. 1983). In itaconic acid-producing A. terreus, however, the canonical view of its biosynthesis assumes that cis-aconitate, formed in the mitochondria, is transported into the cytoplasm and consequently citrate should not accumulate to high amounts there. It is of course possible that cis-aconitate may also inhibit PfkA, but this has not been investigated yet. Itaconate, however, has been reported to be a potent inhibitor of PfkB (forming Fru-2,6-P2; Sakai et al. 2004), and thus the concentration of this major activator of PfkA may become reduced in the presence of itaconate. Since the truncated PfkA has a much higher affinity for Fru-2,6-P2 (Mesojednik and Legiša 2005), this would overcome the shortage of this activator and stimulate the rate of itaconic acid formation.
Are citric acid and itaconic acid completely synthesized in the cytoplasm?
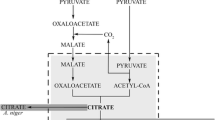
As already described above, the canonical view of biosynthesis of citric and itaconic acid is that the two products of glycolysis—pyruvate and oxaloacetate (the latter formed in the cytosol from pyruvate by carboxylation and transported as malate) enter the mitochondria, and after conversion of pyruvate to acetyl-CoA—are used by citrate synthase to form citric acid. Citric acid is then transported into the cytosol by exchange with malate via the mitochondrial citrate/malate antiporter CtpA. In A. terreus, citrate is first converted to cis-aconitate, which is then exported into the cytosol by the mitochondrial carrier MttA (Hossain et al. 2016). Kirimura et al. (2016) however showed that a ctpA knock-out strain of A. niger still accumulates citric acid with a rate of about 50% of the parental strain. This could be explained by postulating that there may be a second or even more mitochondrial transporters for citric acid. However, the major mitochondrial citrate synthase gene citA is next neighbor to the ctpA in the A. niger genome, and citrate synthase of S. cerevisiae is known to bind CtpA located in the inner mitochondrial membrane (Grigorenko et al. 1990), which suggests that CtpA is the main antiporter transporting citrate into the cytoplasm.
An alternative explanation could be that part of citrate is not synthesized in the mitochondria but in the cytosol. This hypothesis is supported by the presence of two cytosolic isoenzymes of citrate synthase in the cytosol (one being CitB, see above), and by the fact that the overexpression of the mitochondrial citrate synthase CitA in A. niger did not lead to increased citrate accumulation. Since pyruvate decarboxylase is also located in the cytosol, citrate biosynthesis would then only need a cytosolic supply of acetyl-CoA. One possibility, albeit not studied so far, is the oxidation of pyruvate to acetaldehyde and subsequent conversion to acetyl-CoA by the acetyl-CoA synthase FacA. Both genes are present and expressed in A. niger, but have not yet been investigated in this regards. Alternatively, cytosolic acetyl-CoA could be formed by ATP-citrate lyase, whose expression is upregulated in A. niger under high citric acid producing conditions (Chen et al. 2014). This would constitute a futile cycle that converts 1 mol ATP back to ADP and inorganic phosphate. The main benefit of such a mechanism would be the production of less ATP which would be further augmented by alterations in the respiratory chain. The latter aspect and its consequences are described below.
A. terreus has been reported to have no CitB ortholog (Hossain et al. 2016). BLASTp search of its genome, however, retrieved two citrate synthases with moderate similarity to CitB: ATET_2020 (E value 8.3E-43) and ATET_8593 (E value 3.7E-30). Analysis by TargetP shows that ATET_2020 bears a mitochondrial import signal, whereas ATET_8593 does not. Thus the latter is likely located in the cytosol and a cytosolic citrate biosynthesis seems therefore to be possible in A. terreus, too. In fact, when the A. terreus itaconic acid cluster genes were transformed into A. niger, the resulting itaconate formation was stimulated by overexpression of A. niger citB (Hossain et al. 2016). On the other hand, overexpression of the main mitochondrial isoenzyme—CitA—did not result in increased itaconate production (Huang et al. 2014), thus paralleling the results obtained with CitA and citric acid production in A. niger (Ruijter et al. 2000). In addition, A. terreus also has three aconitases of which one is predicted to be localized in the cytosol by TargetP (unpublished data), and cis-aconitate decarboxylase is only found in the cytosol. Furthermore, an acetaldehyde and an acetyl-CoA synthase are—as in A. niger—also present, and a complete cytoplasmic pathway would therefore be possible. It is of interest in this regards that Blumhoff et al. (2013) found that expression and transport of the A. terreus aconitase and cis-aconitate decarboxylase into the same cellular compartment in A. niger leads to higher itaconate production than when they were present in their native compartments. Unfortunately, such investigations have not yet been made with A. terreus so far and the effect of deleting the gene encoding mitochondrial cis-aconitate transporter MttA has not yet been studied either. Its overexpression, however, does not stimulate itaconic acid production, and—in contrast—leads to lower production in some transformants (Huang et al. 2014).
NADH reoxidation and production of ATP
As noted above, the availability of dissolved oxygen at equimolar concentrations to that of glucose is mandatory for citric acid and itaconic acid accumulation in high yields, which is in clear excess of the (minimal) concentration needed for maximal biomass formation (Karaffa and Kubicek 2003; Kubicek and Karaffa 2010). Since co-enzymes such as NAD+ or ATP are present in cells only in limited total amounts, this implies a careful balance of their use and regeneration. The biosynthesis of citrate or itaconate via the canonical pathways produces 2 mol of NADH during glycolysis and a further one during acetyl-CoA formation by the pyruvate dehydrogenase reaction. A strong operation of glycolysis, as occurs during citric acid and itaconic acid fermentation, therefore produces 2 ATP and generates 3 NADH that are converted to additional 6.5 ATP by the operation of the respiratory chain (Table 1). Once growth has stopped, only cell maintenance and pyruvate carboxylase consume ATP, which therefore accumulates and will impair metabolism. It should be noted that the maintenance of the pH gradient between the external medium (pH 1.5) and the cytosol (pH between 6.5 and 7.2; Hesse et al. 2000) also consumes ATP, but the stoichiometry is difficult to calculate because the uptake of nutrients (e.g., glucose) also requires H+ for proton symport.
As a consequence, accumulation of citric and itaconic acid requires alternative mechanisms that are not coupled to ATP generation. One of them is known as the alternative oxidase, which directly oxidizes ubiquinon without proton pumping and consequently without ATP production (Li et al. 2011a, b). It bypasses the cyanide- or antimycin-sensitive steps of the respiratory chain (hence its old name “cyanide-resistant respiration”). It is known to be expressed under conditions of oxidative stress (Abrashev et al. 2008) and thus also during citric and itaconic acid fermentations (Kubicek et al. 1979; Zehentgruber et al. 1980; Kirimura et al. 2016; Molnár et al. 2018). Wallrath et al. (1991) showed that a citric acid-producing mutant strain exhibited reduced activity of the proton-pumping respiratory complexes and the activity of NADH-ubiquinone oxidoreductase was selectively lost at the onset of citric acid production. This loss of NADH-ubiquinon oxidoreductase activity appears to be important for citric acid accumulation because its knock-out resulted in a 20-fold increased concentration of intracellular citric acid, and increased amounts of citrate in the medium (Prömper et al. 1993). Unfortunately, citric acid production reported in this paper was comparably low (40 g/L), and it remains unclear whether these findings would also be valid under overproducing conditions. It nevertheless shows that alternative mechanism for reoxidation of NADH produced in mitochondria must operate during citric acid accumulation.
Wang et al. (2015) extended the above findings by hypothesizing that a reduction in the activity of the later stages of the standard respiratory chain would also stimulate citric acid production under high producing (150–170 g/L) conditions. To this end, they used an inhibitor that blocks ubiquinone-cytochrome C oxidase (antimycin) and the standard uncoupler 2,4-dinitrophenol that dissipates the H+ gradient needed for ATP generation. Both treatments resulted in an about 20% increase in citric acid yield, thus confirming that an interruption of the standard respiratory chain is beneficial also under high citric-producing conditions. These findings would imply that the operation of the alternative oxidase is essential for citric acid accumulation. However, Hou et al. (2018) found that strains in which one of the alternative oxidase genes (aoxA) had been knocked out displayed only a 25% reduced citric acid yield in a high producing strain of A. niger. It must be pointed out, however, that there is also a second alternative oxidase gene in A. niger (aoxB), which has not been studied do far.
Another intriguing aspect of the alternative respiratory chain is that it is rapidly deactivated by short interruptions in the air supply leading to irreversible decreases in citric acid formation (Zehentgruber et al. 1980). This phenomenon was observed in itaconic acid fermentations as well (Gyamerah 1995; Lin et al. 2004; Kuenz et al. 2012). Lin et al. (2004) therefore inserted a hemoglobin gene from the Gram-negative bacterium Vitreoscilla sp. into the A. terreus genome which resulted in a small antagonism against an interruption of aeration under moderate (40 g/L) itaconic acid production.
Regulation of citric acid and itaconic acid accumulation by wide domain regulators
Most of the studies on regulation of formation of these both acids have been done at the level of the metabolic reactions forming and transporting them. No data are so far available whether (and how) they are controlled at a genome wide level. The role of the Zn2Cys6 transcriptional activator encoded in the “itaconate cluster” (see above) would be an attractive candidate. In fact, the “itaconate cluster” genes were identified because of their strong expression in A. terreus (Li et al. 2011a, b), and Kanamasa et al. (2008) reported that cadA is more strongly expressed in a high producer strain of A. terreus, thus pointing to a possible role of the rate of expression in cadA at least. On the other hand, Yin et al. (2017) investigated the transcriptome of two A. niger strains with different productivity during citric acid accumulation and found no significant differences in the expression of genes involved in its biosynthesis and further metabolism. This indicates the necessity for further studies in this area.
The only information concerning a possible wide domain control of citric and itaconic acid relate their overproduction to the protein methyltransferase LaeA (Niu et al. 2015), a protein associated with the conserved transcriptional regulatory velvet complex (Bayram et al. 2008). LaeA has been identified as an activator of secondary metabolite production, but was later also shown to control cellular development and extracellular enzyme formation (Bayram and Braus 2012; Karimi-Aghcheh et al. 2014). Several, but not all LaeA targets have been shown to occur as gene clusters. It would therefore be obvious to explain the effect of LaeA on itaconic acid accumulation on this basis. However, this would not account for the effect on citric acid accumulation: we have checked the genomic location of the genes for citric acid biosynthesis from glucose, and did not find any evidence for their clustering (Supplementary Table S4). Seven cases were found where two genes are in close vicinity, among which are the mitochondrial citrate synthase citA and the tricarboxylic acid transporter ctpA; the gene encoding cytosolic citrate synthase CitB and the one encoding the unknown Zn2Cys6 transcriptional activator that is part of the “itaconate cluster”(see above); and an aconitase with a putative mitochondrial tricarboxylic acid transporter. While we are unaware whether LaeA would specifically act on gene pairs, these data rather suggest that the effect of LaeA on citric acid and itaconic acid biosynthesis cannot be due to an activation of gene clusters synthesizing them. Unfortunately, no gene expression data have yet been published on citric and itaconic acid accumulation in laeA overexpressing and laeA knock-out mutants.
On the other hand, LaeA has been demonstrated to also act on signal transduction and this could be hypothesized to be a reason for its effect on acidogenesis. LaeA has been shown to be necessary for oxidative stress response in A. fumigatus (Fréalle et al. 2013). A role of LaeA in oxidative stress response would be in agreement with its essential role in fungal sporulation (Bayram and Braus 2012), its necessity for extracellular enzyme production (Karimi-Aghcheh et al. 2014), and requirement for secondary metabolite production (Németh et al. 2016). Since both citric and itaconic acid require high oxygen tensions for their accumulation, they are very likely under oxidative stress, although this has never been experimentally investigated so far. Further studies on the role of LaeA on the transcriptome of A. niger and A. terreus under acid-overproducing conditions may therefore contribute significantly to the understanding of these two processes.
Concluding remarks
Although the fundamentals of the fermentation production of citric and itaconic acid have been established for a long while now, and (with the exception of two final steps for itaconic acid formation) the biochemical pathways involved are the same, few attempts have been made for mutual application of this knowledge. Based on the data discussed in this review, there is no need to study the production of citric or itaconic acid under conditions resulting in low titers because both can be produced at high molar yields (Yp/s > 0.7) in their native producers, and there is no reason to assume that A. niger is better suited/adapted for the accumulation of organic acids than A. terreus. This particularly accounts for the attempts to produce itaconic acid heterologously in A. niger: while this may be attractive for reasons of industrial proprietary rights, patents, and regulations, it is still unclear if recombinant strains of A. niger would produce more itaconic acid than the industrial A. terreus strains currently in use, but the data presented in this review make this rather unlikely.
The only differences between citric acid and itaconic acid formation are the two additional steps encoded by mttA and cadA. The two encoded proteins appear to have orthologs in A. niger (and some Penicillium/Talaromyces spp. in the case of cadA), which suggests that they have a special evolutionary origin. Since they occur in cluster with two other genes (a Zn2Cys6 transcription factor and a MFS permease), one would expect that these two genes have a limited distribution, too. In contrast, they are present in a broad range of fungi. Most intriguingly, the A. niger orthologue of the Zn2Cys6 transcription factor also occurs in a gene cluster that encodes enzymes that could have the ability to form tensyuic acid, an alkyl-itaconic acid. The presence of a putative methylcitrate dehydratase (which has no close similarity to CadA), but no CadA in this cluster raises the question whether this methylcitrate dehydratase (and maybe others as well) could form itaconate. In addition, an investigation of the presence of these two clusters in other Aspergillus sps. or Eurotiales may illuminate their origin and evolution.
Understanding the regulation and compartmentation of citric acid and itaconic acid biosynthesis is still insufficient. This warrants more studies with gene deletion mutants that could answer—among others—the following questions: are both citric and itaconic acid regulated by the Zn2Cys6 transcription factor? Are MttA and CitB essential for itaconic acid formation (which would support/reject that hypothesis of a cytosolic biosynthesis)?
Summarizing, a wealth of information is now available about the biosynthesis of citric acid and itaconic acid, but deeper cell biology studies (regulation, compartmentation, energy balance) are still lacking. Such experiments must be performed under conditions of very high productivity because several aspects of cell physiology (compartmentation, gene expression, etc.) may adapt to the intensity of the acid-producing biosynthetic pathways. Finally, if a gene to be investigated is available in both A. niger and A. terreus, we recommend to perform the study in both fungi as the results are likely relevant to the accumulation of both acids.
References
Abrashev RI, Pashova SB, Stefanova LN, Vassilev SV, Dolashka-Angelova PA, Angelova MB (2008) Heat-shock-induced oxidative stress and antioxidant response in Aspergillus niger 26. Can J Microbiol 54:977–983. https://doi.org/10.1139/W08-091
Apelblat A (2014) Citric acid. Springer International Publishing, Switzerland. https://doi.org/10.1007/978-3-319-11233-6_1
Arts E, Kubicek CP, Röhr M (1987) Regulation of phosphofructokinase from Aspergillus niger: effect of fructose 2,6-bisphosphate on the action of citrate, ammonium ions and AMP. Microbiology 133:1195–1199. https://doi.org/10.1099/00221287-133-5-1195
Bafana R, Pandey RA (2018) New approaches for itaconic acid production: bottlenecks and possible remedies. Crit Rev Biotechnol 38:68–82. https://doi.org/10.1080/07388551.2017.1312268
Bambouskova M, Gorvel L, Lampropoulou V, Sergushichev A, Loginicheva E, Johnson K, Korenfeld D, Mathyer ME, Kim H, Huang L-H, Duncan D, Bregman H, Keskin A, Santeford A, Apte RS, Sehgal R, Johnson B, Amarasinghe GK, Soares MP, Satoh T, Akira S, Hai T, de Guzman Strong C, Auclair K, Roddy TP, Biller SA, Jovanovic M, Klechevsky E, Stewart KM, Randolph GJ, Artyomov MN (2018) Electrophilic properties of itaconate and derivatives regulate the IκBζ–ATF3 inflammatory axis. Nature 556:501–504. https://doi.org/10.1038/s41586-018-0052-z
Batti M, Schweiger LB (1963) Process for the production of itaconic acid. US Patent 3078217A
Bayram Ö, Braus GH (2012) Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36:1–24. https://doi.org/10.1111/j.1574-6976.2011.00285.x
Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon N-J, Keller NP, Yu J-H, Braus GH (2008) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320:1504–1506. https://doi.org/10.1126/science.1155888
Bentley R, Thiessen CP (1957) Biosynthesis of itaconic acid in Aspergillus terreus. III. The properties and reaction mechanism of cis-aconitic acid decarboxylase. J Biol Chem 226:703–720
Blatzer M, Latgé JP (2017) Metal-homeostasis in the pathobiology of the opportunistic human fungal pathogen Aspergillus fumigatus. Curr Opin Microbiol 40:152–159
Blumhoff ML, Steiger MG, Mattanovich D, Sauer M (2013) Targeting enzymes to the right compartment: metabolic engineering for itaconic acid production by Aspergillus niger. Metab Eng 19:26–32. https://doi.org/10.1016/j.ymben.2013.05.003
Bonnarme P, Gillet B, Sepulchre AM, Role C, Beloeil JC, Ducrocq C (1995) Itaconate biosynthesis in Aspergillus terreus. J Bacteriol 177:3573–3578. https://doi.org/10.1128/jb.177.12.3573-3578.
Calam CT, Oxford AE, Raistrick H (1939) Studies in the biochemistry of micro-organisms: Itaconic acid, a metabolic product of a strain of Aspergillus terreus Thom. Biochem J 33:1488–1495. https://doi.org/10.1042/bj0331488
Canessa P, Larrondo LF (2013) Environmental responses and the control of iron homeostasis in fungal systems. Appl Microbiol Biotechnol 97:939–955. https://doi.org/10.1007/s00253-012-4615-x
Capuder M, Šolar T, Benčina M, Legiša M (2009) Highly active, citrate inhibition resistant form of Aspergillus niger 6-phosphofructo-1-kinase encoded by a modified pfkA gene. J Biotechnol 144:51–57. https://doi.org/10.1016/j.jbiotec.2009.04.004
Cavallo E, Charreau H, Cerrutti P, Foresti ML (2017) Yarrowia lipolytica: a model yeast for citric acid production. FEMS Yeast Res 17. https://doi.org/10.1093/femsyr/fox084
Chen Y, Nielsen J (2016) Biobased organic acids production by metabolically engineered microorganisms. Curr Opin Biotechnol 37:165–172. https://doi.org/10.1016/J.COPBIO.2015.11.004
Chen H, He X, Geng H, Liu H (2014) Physiological characterization of ATP-citrate lyase in Aspergillus niger. J Ind Microbiol Biotechnol 41:721–731. https://doi.org/10.1007/s10295-014-1418-3
Clark DS (1962) Submerged citric acid fermentation of ferrocyanide-treated cane molasses. Biotechnol Bioeng 4:17–21
Cleland WW, Johnson MJ (1954) Tracer experiments on the mechanism of citric acid formation by Aspergillus niger. J Biol Chem 208:679–689
Cox PW, Paul GC, Thomas CR (1998) Image analysis of the morphology of filamentous micro-organisms. Microbiology 144:817–827. https://doi.org/10.1099/00221287-144-4-817
Currie JN (1917) The citric acid fermentation of Aspergillus niger. J Biol Chem 31:15–37. https://doi.org/10.1200/jco.1993.11.8.1573
Dai Z, Mao X, Magnuson JK, Lasure LL (2004) Identification of genes associated with morphology in Aspergillus niger by using suppression subtractive hybridization. Appl Environ Microbiol 70:2474–2485
Doelger WP, Prescott SC (1934) Citric acid fermentation. Ind Eng Chem 26:1142–1149. https://doi.org/10.1021/ie50299a005
El-Imam AA, Du C (2014) Biodiversity, bioprospecting and development. J Biodivers Biopros Dev 1:1–8. https://doi.org/10.4172/ijbbd.1000119
Entian KD, Barnett JA (1992) Regulation of sugar utilization by Saccharomyces cerevisiae. Trends Biochem Sci 17:506–510. https://doi.org/10.1016/0968-0004(92)90341-6
Fiechter A, Gmünder FK (1989) Metabolic control of glucose degradation in yeast and tumor cells. Adv Biochem Eng Biotechnol 39:1–28. https://doi.org/10.1007/BFb0051950
Foster JW (1949) Chemical activities of Fungi. Academic Press, New York
Fréalle E, Aliouat-Denis C-M, Delhaes L, Hot D, Dei-Cas E (2013) Transcriptomic insights into the oxidative response of stress-exposed Aspergillus fumigatus. Curr Pharm Des 19:3713–3737. https://doi.org/10.2174/1381612811319200011
Geiser E, Przybilla SK, Friedrich A, Buckel W, Wierckx N, Blank LM, Bölker M (2016) Ustilago maydis produces itaconic acid via the unusual intermediate trans -aconitate. Microb Biotechnol 9:116–126. https://doi.org/10.1111/1751-7915.12329
Gil-Bona A, Llama-Palacios A, Parra CM, Vivanco F, Nombela C, Monteoliva L, Gil C (2015) Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J Proteome Res 14:142–153. https://doi.org/10.1021/pr5007944
Grigorenko EV, Small WC, Persson L-O, Srere PA (1990) Citrate synthase 1 interacts with the citrate transporter of yeast mitochondria. J Mol Recognit 3:215–219. https://doi.org/10.1002/jmr.300030508
Guerinot ML, Meidl EJ, Plessner O (1990) Citrate as a siderophore in Bradyrhizobium japonicum. J Bacteriol 172:3298–3303
Guevarra ED, Tabuchi T (1990) Accumulation of itaconic, 2-hydroxyparaconic, itatartaric, and malic acids by strains of the genus Ustilago. J Agric Biol Chem 54:2353–2358. https://doi.org/10.1080/00021369.1990.10870333
Gyamerah M (1995) Factors affecting the growth form of Aspergillus terreus NRRL 1960 in relation to itaconic acid fermentation. Appl Microbiol Biotechnol 44:356–361. https://doi.org/10.1007/BF00169929
Habison A, Kubicek CP, Röhr M (1983) Partial purification and regulatory properties of phosphofructokinase from Aspergillus niger. Biochem J 209:669–676. https://doi.org/10.1042/BJ2090669
Hasegawa Y, Fukuda T, Hagimori K, Tomoda H, Omura S (2007) Tensyuic acids, new antibiotics produced by Aspergillus niger FKI-2342. Chem Pharm Bull 55:1338–1341
Hesse SJA, Ruijter GJG, Dijkema C, Visser J (2000) Measurement of intracellular (compartmental) pH by 31P NMR in Aspergillus niger. J Biotechnol 77:5–15
Hevekerl A, Kuenz A, Vorlop K-D (2014) Influence of the pH on the itaconic acid production with Aspergillus terreus. Appl Microbiol Biotechnol 98:10005–10012
Hossain AH, Li A, Brickwedde A, Wilms L, Caspers M, Overkamp K, Punt PJ (2016) Rewiring a secondary metabolite pathway towards itaconic acid production in Aspergillus niger. Microb Cell Factories 15:130
Hou L, Liu L, Zhang H, Zhang L, Zhang L, Zhang J, Gao Q, Wang D (2018) Functional analysis of the mitochondrial alternative oxidase gene (aox1) from Aspergillus niger CGMCC 10142 and its effects on citric acid production. Appl Microbiol Biotechnol 102:7981–7995. https://doi.org/10.1007/s00253-018-9197-9
Huang X, Lu X, Li Y, Li X, Li J-J (2014) Improving itaconic acid production through genetic engineering of an industrial Aspergillus terreus strain. Microb Cell Factories 13:119. https://doi.org/10.1186/s12934-014-0119-y
Kanamasa S, Dwiarti L, Okabe M, Park EY (2008) Cloning and functional characterization of the cis-aconitic acid decarboxylase (CAD) gene from Aspergillus terreus. Appl Microbiol Biotechnol 80:223–229
Kane JA, Finlay AC, Amann PF (1945) Production of itaconic acid. US Patent 2385283A
Karaffa L, Kubicek CP (2003) Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl Microbiol Biotechnol 61:189–196. https://doi.org/10.1007/s00253-002-1201-7
Karaffa L, Díaz R, Papp B, Fekete E, Sándor E, Kubicek CP (2015) A deficiency of manganese ions in the presence of high sugar concentrations is the critical parameter for achieving high yields of itaconic acid by Aspergillus terreus. Appl Microbiol Biotechnol 99:7937–7944. https://doi.org/10.1007/s00253-015-6735-6
Karimi-Aghcheh R, Németh Z, Atanasova L, Fekete E, Paholcsek M, Sándor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP (2014) The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS One 9:e112799. https://doi.org/10.1371/journal.pone.0112799
Kinoshita K (1931) Production of itaconic acid and mannitol by a new mold, Aspergillus itaconicus. Acta Phytochim (Japan) 5:271–287
Kirimura K, Kobayashi K, Ueda Y, Hattori T (2016) Phenotypes of gene disruptants in relation to a putative mitochondrial malate–citrate shuttle protein in citric acid-producing Aspergillus niger. Biosci Biotechnol Biochem 80:1737–1746. https://doi.org/10.1080/09168451.2016.1164583
Kisser M, Kubicek CP, Röhr M (1980) Influence of manganese on morphology and cell wall composition of Aspergillus niger during citric acid fermentation. Arch Microbiol 128:26–33. https://doi.org/10.1007/BF00422301
Kobayashi E, Suzuki T, Yamamoto M (2013) Roles nrf2 plays in myeloid cells and related disorders. Oxidative Med Cell Longev 2013:529219–529217. https://doi.org/10.1155/2013/529219
Krebs HA (1940) The citric acid cycle and the Szent-Györgyi cycle in pigeon breast muscle. Biochem J 34:775–779. https://doi.org/10.1042/bj0340775
Kubicek CP, Karaffa L (2010) Citric acid processes. In: Flickinger MC (ed) Encyclopedia of industrial biotechnology: bioprocess, bioseparation, and cell technology, vol 3. John Wiley & Sons, Inc., Hoboken, pp 1652–1658
Kubicek CP, Röhr M (1985) Aconitase and citric acid fermentation by Aspergillus niger. Appl Environ Microbiol 50:1336–1338. https://doi.org/10.1007/BF00422301
Kubicek CP, Hampel W, Röhr M (1979) Manganese deficiency leads to elevated amino acid pools in citric acid accumulating Aspergillus niger. Arch Microbiol 123:73–79. https://doi.org/10.1007/BF00403504
Kubicek CP, Hönlinger C, Jaklitsch WM, Affenzeller K, Mach R, Gerngross T-U, Ying L (1990) Regulation of lysine biosynthesis in the fungus Penicillium chrysogenum. In: Amino Acids. Springer Netherlands, Dordrecht, pp 1029–1034. https://doi.org/10.1007/978-94-011-2262-7_128
Kuenz A, Krull S (2018) Biotechnological production of itaconic acid—things you have to know. Appl Microbiol Biotechnol 102:3901–3914. https://doi.org/10.1007/s00253-018-8895-7
Kuenz A, Gallenmüller Y, Willke T, Vorlop K-D (2012) Microbial production of itaconic acid: developing a stable platform for high product concentrations. Appl Microbiol Biotechnol 96:1209–1216. https://doi.org/10.1007/s00253-012-4221-y
Li Q, Bai Z, O'Donnell A, Harvey LM, Hoskisson PA, McNeil B (2011a) Oxidative stress in fungal fermentation processes: the roles of alternative respiration. Biotechnol Lett 33:457–467
Li A, van Luijk N, ter Beek M, Caspers M, Punt P, van der Werf M (2011b) A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus. Fungal Genet Biol 48:602–611
Lin Y-H, Li Y-F, Huang M-C, Tsai Y-C (2004) Intracellular expression of Vitreoscilla hemoglobin in Aspergillus terreus to alleviate the effect of a short break in aeration during culture. Biotechnol Lett 26:1067–1072. https://doi.org/10.1023/B:BILE.0000032964.15178.7c
Lockwood LB, Nelson GE (1946) Some factors affecting the production of itaconic acid by Aspergillus terreus in agitated cultures. Arch Biochem 10:365–374
Martin SM, Waters WR (1952) Production of citric acid by submerged fermentation. Ind Eng Chem 44:2229–2233. https://doi.org/10.1021/ie50513a067
McFadden BA, Williams JO, Roche TE (1971) Mechanism of action of isocitrate lyase from Pseudomonas indigofera. Biochemistry 10:1384–1390. https://doi.org/10.1021/bi00784a017
Mesojednik S, Legiša M (2005) Posttranslational modification of 6-phosphofructo-1-kinase in Aspergillus niger. Appl Environ Microbiol 71:1425–1432. https://doi.org/10.1128/AEM.71.3.1425-1432.2005
Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, Buttini M, Linster CL, Medina E, Balling R, Hiller K (2013) Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci U S A 110:7820–7825. https://doi.org/10.1073/pnas.1218599110
Mlakar T, Legiša M (2006) Citrate inhibition-resistant form of 6-phosphofructo-1-kinase from Aspergillus niger. Appl Environ Microbiol 72:4515–4521. https://doi.org/10.1128/AEM.00539-06
Molnár ÁP, Németh Z, Kolláth IS, Fekete E, Flipphi M, Ág N, Soós Á, Kovács B, Sándor E, Kubicek CP, Karaffa L (2018) High oxygen tension increases itaconic acid accumulation, glucose consumption, and the expression and activity of alternative oxidase in Aspergillus terreus. Appl Microbiol Biotechnol 102:8799–8808. https://doi.org/10.1007/s00253-018-9325-6
Németh Z, Molnár ÁP, Fejes B, Novák L, Karaffa L, Keller NP, Fekete E (2016) Growth-phase sterigmatocystin formation on lactose is mediated via low specific growth rates in Aspergillus nidulans. Toxins 8:354
Niu J, Arentshorst M, Nair PDS, Dai Z, Baker SE, Frisvad JC, Nielsen KF, Punt PJ, Ram AFJ (2015) Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 genes, genomes. Genet 6:193–204. https://doi.org/10.1534/g3.115.024067
Nubel RC, Ratajak EJ (1962) Process for producing itaconic acid. US Patent 3,044,941
Odoni DI, van Gaal MP, Schonewille T, Tamayo-Ramos JA, Martins Dos Santos VAP, Suarez-Diez M, Schaap PJ (2017) Aspergillus niger secretes citrate to increase iron bioavailability. Front Microbiol 8:1424
Okabe M, Lies D, Kanamasa S, Park EY (2009) Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl Microbiol Biotechnol 84:597–606. https://doi.org/10.1007/s00253-009-2132-3
Papagianni M, Mattey M (2006) Morphological development of Aspergillus niger in submerged citric acid fermentation as a function of the spore inoculum level. Application of neural network and cluster analysis for characterization of mycelial morphology. Microb Cell Factories 5(3):3. https://doi.org/10.1186/1475-2859-5-3
Pardo M, Monteoliva L, Vázquez P, Martínez R, Molero G, Nombela C, Gil C (2004) PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 150:4157–4170. https://doi.org/10.1099/mic.0.26924-0
Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck PWM, van Dijk A, Dijkhuizen L, Driessen AJM, D’Enfert C, Geysens S, Goosen C, Groot GSP, de Groot PWJ, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JPTW, van den Hondel CAMJJ, van der Heijden RTJM, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJEC, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NNME, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJJ, Wedler H, Wösten HAB, Zeng A-P, van Ooyen AJJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231. https://doi.org/10.1038/nbt1282
Perlman D, Dorrell WW, Johnson MJ (1946a) Effect of metallic ions on the production of citric acid by Aspergillus niger. Arch Biochem 11:131–143
Perlman D, Kita DA, Peterson WH (1946b) Production of citric acid from cane molasses. Arch Biochem 11:123–129
Prömper C, Schneider R, Weiss H (1993) The role of the proton-pumping and alternative respiratory chain NADH:ubiquinone oxidoreductases in overflow catabolism of Aspergillus niger. Eur J Biochem 216:223–230. https://doi.org/10.1111/j.1432-1033.1993.tb18136.x
Ramakrishnan CV, Steel R, Lentz CP (1955) Mechanism of citric acid formation and accumulation in Aspergillus niger. Arch Biochem Biophys 55:270–273
Ruijter GJ, Panneman H, Xu D, Visser J (2000) Properties of Aspergillus niger citrate synthase and effects of citA overexpression on citric acid production. FEMS Microbiol Lett 184:35–40
Saha BC, Kennedy GJ, Bowman MJ, Qureshi N, Dunn RO (2018) Factors affecting production of Itaconic acid from mixed sugars by Aspergillus terreus. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-018-2831-2
Sakai A, Kusumoto A, Kiso Y, Furuya E (2004) Itaconate reduces visceral fat by inhibiting fructose 2,6-bisphosphate synthesis in rat liver. Nutrition 20:997–1002. https://doi.org/10.1016/j.nut.2004.08.007
Salas ML, Vinuela E, Salas M, Sols A (1965) Citrate inhibition of phosphofructokinase and the pasteur effect. Biochem Biophys Res Commun 19:371–376. https://doi.org/10.1016/0006-291X(65)90471-7
Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA (2014) Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10:371–377. https://doi.org/10.1038/nchembio.1482
Shin WS, Lee D, Kim S, Jeong YS, Chun GT (2013) Application of scale-up criterion of constant oxygen mass transfer coefficient (k L a) for production of itaconic acid in a 50 L pilot-scale fermentor by fungal cells of Aspergillus terreus. J Microbiol Biotechnol 23:1445–1453
Shin WS, Park B, Lee D, Oh MK, Chun GT, Kim S (2017) Enhanced production of itaconic acid through development of transformed fungal strains of Aspergillus terreus. J Microbiol Biotechnol 27:306–315
Snell RL, Schweiger LB (1949) Production of citric acid by fermentation. US Patent 2,492,667
Steel R, Lentz CP, Martin SM (1955) Submerged citric acid fermentation of sugar beet molasses: increase in scale. Can J Microbiol 1:299–311. https://doi.org/10.1139/m55-042
Steiger MG, Blumhoff ML, Mattanovich D, Sauer M (2013) Biochemistry of microbial itaconic acid production. Front Microbiol 4:23. https://doi.org/10.3389/fmicb.2013.00023
Strelko CL, Lu W, Dufort FJ, Seyfried TN, Chiles TC, Rabinowitz JD, Roberts MF (2011) Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc 133:16386–16389. https://doi.org/10.1021/ja2070889
Sun X, Wu H, Zhao G, Li Z, Wu X, Liu H, Zheng Z (2018) Morphological regulation of Aspergillus niger to improve citric acid production by chsC gene silencing. Bioprocess Biosyst Eng 41:1029–1038. https://doi.org/10.1007/s00449-018-1932-1
Takahashi J, Yamada K (1959) Studies on the effects of some physical conditions on the submerged Mold culture. Part II. J Agric Chem Soc Jpn 33:707–710. https://doi.org/10.1271/nogeikagaku1924.33.8_707
Takahashi J, Yamada K, Asai T (1958) Studies on the effects of some physical conditions on the submerged mold culture. J Agric Chem Soc Jpn 32:501–506. https://doi.org/10.1271/nogeikagaku1924.32.7_501
Tevz G, Bencina M, Legisa M (2010) Enhancing itaconic acid production by Aspergillus terreus. Appl Microbiol Biotechnol 87:1657–1664. https://doi.org/10.1007/s00253-010-2642-z
Tomlinson N, Campbell JJR, Trussellb PC (1950) The influence of zinc, iron, copper, and manganese on the production of citric acid by Aspergillus niger. J Bacteriol 59:217–227
van der Straat L, de Graaff LH (2014) Pathway transfer in fungi. Bioengineered 5:335–339. https://doi.org/10.4161/bioe.29936
van der Straat L, Vernooij M, Lammers M, van den Berg W, Schonewille T, Cordewener J, van der Meer I, Koops A, de Graaff LH (2014) Expression of the Aspergillus terreus itaconic acid biosynthesis cluster in Aspergillus niger. Microb Cell Factories 13:11. https://doi.org/10.1186/1475-2859-13-11
van Steenwyk G, Roszkowski M, Manuella F, Franklin TB, Mansuy IM (2018) Transgenerational inheritance of behavioral and metabolic effects of paternal exposure to traumatic stress in early postnatal life: evidence in the 4th generation. Environ Epigenet 4:dvy023. https://doi.org/10.1093/eep/dvy023
Vesth TC, Nybo JL, Theobald S, Frisvad JC, Larsen TO, Nielsen KF, Hoof JB, Brandl J, Salamov A, Riley R, Gladden JM, Phatale P, Nielsen MT, Lyhne EK, Kogle ME, Strasser K, McDonnell E, Barry K, Clum A, Chen C, LaButti K, Haridas S, Nolan M, Sandor L, Kuo A, Lipzen A, Hainaut M, Drula E, Tsang A, Magnuson JK, Henrissat B, Wiebenga A, Simmons BA, Mäkelä MR, de Vries RP, Grigoriev IV, Mortensen UH, Baker SE, Andersen MR (2018) Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat Genet 50:1688–1695. https://doi.org/10.1038/s41588-018-0246-1
Wallrath J, Schmidt M, Weiss H (1991) Correlation between manganese-deficiency, loss of respiratory chain complex I activity and citric acid production in Aspergillus niger. Arch Microbiol 158:435–438. https://doi.org/10.1007/BF00276305
Wang L, Zhang J, Cao Z, Wang Y, Gao Q, Zhang J, Wang D (2015) Inhibition of oxidative phosphorylation for enhancing citric acid production by Aspergillus niger. Microb Cell Factories 14:7. https://doi.org/10.1186/s12934-015-0190-z
Werpy T, Petersen G (2004) Top value added chemicals from biomass: Volume I: results of screening for potential candidates from sugars and synthesis gas. https://doi.org/10.2172/15008859
Yan N (2013) Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci 38:151–159. https://doi.org/10.1016/j.tibs.2013.01.003
Yin X, Shin H, Li J, Du G, Liu L, Chen J (2017) Comparative genomics and transcriptome analysis of Aspergillus niger and metabolic engineering for citrate production. Sci Rep 7:41040. https://doi.org/10.1038/srep41040
Yu B, Zhang X, Sun W, Xi X, Zhao N, Huang Z, Ying Z, Liu L, Liu D, Niu H, Wu J, Zhuang W, Zhu C, Chen Y, Ying H (2018) Continuous citric acid production in repeated-fed batch fermentation by Aspergillus niger immobilized on a new porous foam. J Biotechnol 276-277:1–9. https://doi.org/10.1016/j.jbiotec.2018.03.015
Zehentgruber O, Kubicek CP, Röhr M (1980) Alternative respiration of Aspergillus niger. FEMS Microbiol Lett 8:71–74. https://doi.org/10.1111/j.1574-6968.1980.tb05052.x
Zhao M, Lu X, Zong H, Li J, Zhuge B (2018) Itaconic acid production in microorganisms. Biotechnol Lett 40:455–464. https://doi.org/10.1007/s10529-017-2500-5
Acknowledgements
Research in LK’s lab is supported by the EU and co-financed by the European Regional Development Fund under the project GINOP-2.3.2-15-2016-00008; by the EFOP-3.6.1-16-2016-00022 project co-financed by the European Union and the European Social Fund; and by the National Research, Development and Innovation Fund (KH 129602, NN128867 and NN116519). The authors are grateful to Erzsébet Fekete, Norbert Ág, István S. Kolláth, and Balázs Fejes (all at the University of Debrecen) for their help.
Funding
Open access funding provided by University of Debrecen (DE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 4960 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Karaffa, L., Kubicek, C.P. Citric acid and itaconic acid accumulation: variations of the same story?. Appl Microbiol Biotechnol 103, 2889–2902 (2019). https://doi.org/10.1007/s00253-018-09607-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-09607-9