Abstract
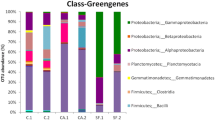
Comprehensive studies of the biodiversity of the microbial epilithic community on monuments may provide critical insights for clarifying factors involved in the colonization processes. We carried out a high-throughput investigation of the communities colonizing the medieval church of San Leonardo di Siponto (Italy) by Illumina-based deep sequencing. The metagenomic analysis of sequences revealed the presence of Archaea, Bacteria, and Eukarya. Bacteria were Actinobacteria, Proteobacteria, Bacteroidetes, Cyanobacteria, Chloroflexi, Firmicutes and Candidatus Saccharibacteria. The predominant phylum was Actinobacteria, with the orders Actynomycetales and Rubrobacteriales, represented by the genera Pseudokineococcus, Sporichthya, Blastococcus, Arthrobacter, Geodermatophilus, Friedmanniella, Modestobacter, and Rubrobacter, respectively. Cyanobacteria sequences showing strong similarity with an uncultured bacterium sequence were identified. The presence of the green algae Oocystaceae and Trebuxiaceae was revealed. The microbial diversity was explored at qualitative and quantitative levels, evaluating the richness (the number of operational taxonomic units (OTUs)) and the abundance of reads associated with each OTU. The rarefaction curves approached saturation, suggesting that the majority of OTUs were recovered. The results highlighted a structured community, showing low diversity, made up of extremophile organisms adapted to desiccation and UV radiation. Notably, the microbiome appeared to be composed not only of microorganisms possibly involved in biodeterioration but also of carbonatogenic bacteria, such as those belonging to the genus Arthrobacter, which could be useful in bioconservation. Our investigation demonstrated that molecular tools, and in particular the easy-to-run next-generation sequencing, are powerful to perform a microbiological diagnosis in order to plan restoration and protection strategies.





Similar content being viewed by others
References
Andriani G, Walsh N (2003) Fabric, porosity and water permeability of calcarenites from Apulia (SE Italy) used as building and ornamental stone. Bull Eng Geol Environ 62:77–84
Barnard RL, Osborne CA, Firestone MK (2013) Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J 7(11):2229–2241. doi:10.1038/ismej.2013.104
Berdoulay M, Salvado JC (2009) Genetic characterization of microbial communities living at the surface of building stones. Lett Appl Microbiol 49(3):311–316. doi:10.1111/j.1472-765X.2009.02660.x
Chakravorty S, Helb D, Burday M, Connell N, Alland D (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69(2):330–339
Dakal TC, Cameotra SS (2012) Microbially induced deterioration of architectural heritages: routes and mechanisms involved. Environmental Sciences Europe 24:36
de Castro AP, Sartori da Silva MR, Quirino BF, Kruger RH (2013) Combining “omics” strategies to analyze the biotechnological potential of complex microbial environments. Curr Protein Pept Sci 14(6):447–458
de Wit R, Bouvier T (2006) ‘Everything is everywhere, but, the environment selects’; what did baas Becking and Beijerinck really say? Environ Microbiol 8(4):755–758
Decelle J, Romac S, Stern RF, Bendif el M, Zingone A, Audic S, Guiry MD, Guillou L, Tessier D, Le Gall F, Gourvil P, Dos Santos AL, Probert I, Vaulot D, de Vargas C, Christen R (2015) PhytoREF: a reference database of the plastidial 16S rRNA gene of photosynthetic eukaryotes with curated taxonomy. Mol Ecol Resour 15(6):1435–1445. doi:10.1111/1755-0998.12401
Ditaranto N, Loperfido S, van der Werf I, Mangone A, Cioffi N, Sabbatini L (2011) Synthesis and analytical characterisation of copper-based nanocoatings for bioactive stone artworks treatment. Anal Bioanal Chem 399:473–481. doi:10.1007/s00216-010-4301-8
Ditaranto N, van der Werf ID, Picca RA, Sportelli MC, Giannossa LC, Bonerba E, Tantillo G, Sabbatini L (2015) Characterization and behaviour of ZnO-based nanocomposites designed for the control of biodeterioration of patrimonial stoneworks. New J Chem 39:6836–6843. doi:10.1039/C5NJ00527B
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. doi:10.1093/bioinformatics/btr381
Ettenauer JD, Jurado V, Piñar G, Miller AZ, Santner M, Saiz-Jimenez C, Sterflinger K (2014) Halophilic microorganisms are responsible for the rosy discolouration of saline environments in three historical buildings with mural paintings. PLoS One 9(8):e103844. doi:10.1371/journal.pone.0103844
Friedmann EI, Ocampo-Friedmann R (1984) The Antarctic cryptoendolithic ecosystem: relevance to exobiology. Orig Life 14(1–4):771–776
Guillitte O (1995) Bioreceptivity: a new concept for building ecology studies. Sci Total Environ 167:215–220
Gutarowska B, Celikkol-Aydin S, Bonifay V, Otlewska A, Aydin E, Oldham AL, Brauer JI, Duncan KE, Adamiak J, Sunner JA, Beech IB (2015) Metabolomic and high-throughput sequencing analysis-modern approach for the assessment of biodeterioration of materials from historic buildings. Front Microbiol 6:979. doi:10.3389/fmicb.2015.00979
Hallmann C, Stannek L, Fritzlar D, Hause-Reitner D, Friedl T, Hoppert M (2013) Molecular diversity of phototrophic biofilms on building stone. FEMS Microbiol Ecol 84(2):355–372. doi:10.1111/1574-6941.12065
Hueck HJ (2001) The biodeterioration of materials—an appraisal. Int Biodeterior Biodegrad 48:5–11
Jroundi F, Fernández-Vivas A, Rodriguez-Navarro C, Bedmar EJ, González-Muñoz MT (2010) Bioconservation of deteriorated monumental calcarenite stone and identification of bacteria with carbonatogenic activity. Microb Ecol 60(1):39–54. doi:10.1007/s00248-010-9665-y
Jroundi F, Gómez-Suaga P, Jimenez-Lopez C, González-Muñoz MT, Fernandez-Vivas MA (2012) Stone-isolated carbonatogenic bacteria as inoculants in bioconsolidation treatments for historical limestone. Sci Total Environ 425:89–98. doi:10.1016/j.scitotenv.2012.02.059
Jroundi F, Gonzalez-Muñoz MT, Sterflinger K, Piñar G (2015) Molecular tools for monitoring the ecological sustainability of a stone bio-consolidation treatment at the Royal Chapel, Granada. PLoS One 10(7):e0132465. doi:10.1371/journal.pone.0132465
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79(17):5112–5120. doi:10.1128/AEM.01043-13
Kuhlman KR, Fusco WG, La Duc MT, Allenbach LB, Ball CL, Kuhlman GM, Anderson RC, Erickson IK, Stuecker T, Benardini J, Strap JL, Crawford RL (2006) Diversity of microorganisms within rock varnish in the Whipple Mountains, California. Appl Environ Microbiol 72(2):1708–1715. doi:10.1128/AEM.72.2.1708-1715.2006
Laiz L, Miller AZ, Jurado V, Akatova E, Sanchez-Moral S, Gonzalez JM, Dionísio A, Macedo MF, Saiz-Jimenez C (2009) Isolation of five Rubrobacter strains from biodeteriorated monuments. Naturwissenschaften 96(1):71–79. doi:10.1007/s00114-008-0452-2
Macedo MF, Miller AZ, Dionísio A, Saiz-Jimenez C (2009) Biodiversity of cyanobacteria and green algae on monuments in the Mediterranean Basin: an overview. Microbiology 155(Pt 11):3476–3490. doi:10.1099/mic.0.032508-0
Mann S (2001) Biomineralization: principles and concepts in bioinorganic materials chemistry. Oxford University Press, Oxford, United Kingdom
Manzari C, Fosso B, Marzano M, Annese A, Caprioli R, D’Erchia AM, Gissi C, Intranuovo M, Picardi E, Santamaria M, Scorrano S, Sgaramella G, Stabili L, Piraino S, Pesole G (2015) The influence of invasive jellyfish blooms in a coastal lagoon (Varano, SE Italy) detected by an Illumina-based deep sequencing strategy. Biol Invasions 17:923–940. doi:10.1007/s10530-014-0810-2
Modi SR, Collins JJ, Relman DA (2014) Antibiotics and the gut microbiota. J Clin Invest 124(10):4212–4218. doi:10.1172/JCI72333
Mohammadipanah F, Wink J (2016) Actinobacteria from arid and desert habitats: diversity and biological activity. Front Microbiol 6:1541. doi:10.3389/fmicb.2015.01541
Nesme J, Simonet P (2015) The soil resistome: a critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ Microbiol 17(4):913–930. doi:10.1111/1462-2920.12631
Oksanen J, Blanchet FG, Kindt R, Legendre P, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Vegan: community ecology package. R Packag Version 2:7
Ortega-Calvo JJ, Hernandez-Marine M, Saiz-Jimenez C (1993) Cyanobacteria and algae on historic buildings and monuments. In Naya Prokash (ed) Recent advances in biodeterioration and degradation 1: biodeterioration of cultural heritage, pp 173–203
Piñar G, Ripka K, Weber J, Sterflinger K (2009) The micro-biota of a sub-surface monument the medieval chapel of St. Virgil (Vienna, Austria). Int Biodeterior Biodegrad 63:851–859. doi:10.1016/j.ibiod.2009.02.004
Pinna D (2014) Biofilms and lichens on stone monuments: do they damage or protect? Front Microbiol 5:133. doi:10.3389/fmicb.2014.00133
Portillo MC, Alloza R, Gonzales JM (2009) Three different phototrophic microbial communities colonizing a single natural shelter containing prehistoric paintings. Sci Total Environ 407(17):4876–4881. doi:10.1016/j.scitotenv.2009.05.038
Ragon M, Restoux G, Moreira D, Møller AP, López-García P (2011) Sunlight-exposed biofilm microbial communities are naturally resistant to Chernobyl ionizing-radiation levels. PLoS One 6(7):e21764. doi:10.1371/journal.pone.0021764
Ragon M, Fontaine MC, Moreira D, López-García P (2012) Different biogeographic patterns of prokaryotes and microbial eukaryotes in epilithic biofilms. Mol Ecol 21(15):3852–3868. doi:10.1111/j.1365-294X.2012.05659.x
Santhosh K, Ramachandran SK, Ramakrishnan V, Bang SS (2001) Remediation of concrete using microrganisms. Am Conc Inst Mater J 98:3–9
Scheerer S, Ortega-Morales O, Gaylarde C (2009) Microbial deterioration of stone monuments—an updated overview. Adv Appl Microbiol 66:97–139. doi:10.1016/S0065-2164(08)00805-8
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541. doi:10.1128/AEM.01541-09
Sghaier H, Hezbri K, Ghodhbane-Gtari F, Pujic P, Sen A, Daffonchio D, Boudabous A, Tisa LS, Klenk HP, Armengaud J, Normand P, Gtari M (2016) Stone-dwelling actinobacteria Blastococcus saxobsidens, Modestobacter marinus and Geodermatophilus obscurus proteogenomes. ISME J 10(1):21–29. doi:10.1038/ismej.2015.108
Smith DP, Peay KG (2014) Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS One 9(2):e90234. doi:10.1371/journal.pone.0090234
Starke V, Kirshtein J, Fogel ML, Steele A (2013) Microbial community composition and endolith colonization at an Arctic thermal spring are driven by calcite precipitation. Environ Microbiol Rep 5(5):648–659. doi:10.1111/1758-2229.12063
Sterflinger K, Piñar G (2013) Microbial deterioration of cultural heritage and works of art—tilting at windmills? Appl Microbiol Biotechnol 97(22):9637–9646. doi:10.1007/s00253-013-5283
Suihko ML, Alakomi HL, Gorbushina A, Fortune I, Marquardt J, Saarela M (2007) Characterization of aerobic bacterial and fungal microbiota on surfaces of historic Scottish monuments. Syst Appl Microbiol 30(6):494–508
Tiano P, Accolla P, Tomaselli L (1995) Phototrophic biodeteriogens on lithoid surfaces: an ecological study. Microb Ecol 29(3):299–309. doi:10.1007/BF00164892
Tringe SG, Hugenholtz P (2008) A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 11(5):442–446. doi:10.1016/j.mib.2008.09.011
Urzì C, Realini M (1998) Colour changes of Noto’s calcareous sandstone as related to its colonisation by microorganisms. Int Biodeterior Biodegrad 42:45–54. doi:10.1016/S0964-8305(98)00045-6
Urzì C, Brusetti L, Salamone P, Sorlini C, Stackebrandt E, Daffonchio D (2001) Biodiversity of Geodermatophilaceae isolated from altered stones and monuments in the Mediterranean basin. Environ Microbiol 3(7):471–479
van der Werf ID, Ditaranto N, Picca RA, Sportelli MC, Sabbatini L (2015) Development of a novel conservation treatment of stone monuments with bioactive nanocomposites. Heritag Sci 3:29. doi:10.1186/s40494-015-0060-3
Walker JJ, Pace NR (2007) Phylogenetic composition of Rocky Mountain endolithic microbial ecosystems. Appl Environ Microbiol 73(11):3497–3504
Ward DM, Weller R, Bateson MM (1990) 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345(6270):63–65
Warscheid T, Braams J (2000) Biodeterioration of stone: a review. Int Biodeterior Biodegrad 46(4):343–368. doi:10.1016/S0964-8305(00)00109-8
Acknowledgments
The authors would like to thank Prof. P. Acquafredda (Dept. Scienze della Terra e Geoambientali, University of Bari “Aldo Moro”) for the optical microscopy study of the stone samples and Architect Antonello D’Ardes (Manfredonia, Italy) for providing logistic support. This work was performed in the framework of the PRIN 2010–2011 “Sustainability in cultural heritage: from diagnosis to the development of innovative systems for consolidation, cleaning and protection” code 2010329WPF, funded by MIUR (Italy) and used the facilities of the Molecular Biodiversity Laboratory (MoBiLab) of LifeWatch-Italy. I.D. van der Werf is supported by the Future in Research program financed by the Apulian Region (Italy).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 367 kb)
Rights and permissions
About this article
Cite this article
Chimienti, G., Piredda, R., Pepe, G. et al. Profile of microbial communities on carbonate stones of the medieval church of San Leonardo di Siponto (Italy) by Illumina-based deep sequencing. Appl Microbiol Biotechnol 100, 8537–8548 (2016). https://doi.org/10.1007/s00253-016-7656-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7656-8