Abstract
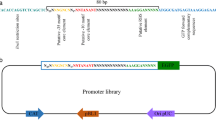
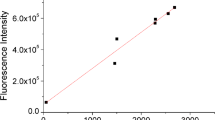
In recent years, the application of transcription factor-based biosensors for the engineering of microbial production strains opened up new opportunities for industrial biotechnology. However, the design of synthetic regulatory circuits depends on the selection of suitable transcription factor-promoter pairs to convert the concentration of effector molecules into a measureable output. Here, we present an efficient strategy to screen promoter libraries for appropriate parts for biosensor design. To this end, we pooled the strains of the Alon library containing about 2000 different Escherichia coli promoter-gfpmut2 fusions, and enriched galactose- and l-phenylalanine-responsive promoters by toggled rounds of positive and negative selection using fluorescence-activated cell sorting (FACS). For both effectors, responsive promoters were isolated and verified by cultivation in microtiter plates. The promoter of mtr, encoding an l-tryptophan-specific transporter, was identified as suitable part for the construction of an l-phenylalanine biosensor. In the following, we performed a comparative analysis of different biosensor constructs based on the mtr promoter. The obtained data revealed a strong influence of the biosensor architecture on the performance characteristics. For proof-of-principle, the mtr sensor was applied in a FACS high-throughput screening of an E. coli MG1655 mutant library for the isolation of l-phenylalanine producers. These results emphasize the developed screening approach as a convenient strategy for the identification of effector-responsive promoters for the design of novel biosensors.




Similar content being viewed by others
References
Backman K, O'Connor MJ, Maruya A, Rudd E, McKay D, Balakrishnan R, Radjai M, DiPasquantonio V, Shoda D, Hatch R, Venkatasubramanian K (1990) Genetic engineering of metabolic pathways applied to the production of phenylalanine. Ann N Y Acad Sci 589:16–24. doi:10.1111/j.1749-6632.1990.tb24231.x
Bang HB, Lee YH, Kim SC, Sung CK, Jeong KJ (2016) Metabolic engineering of Escherichia coli for the production of cinnamaldehyde. Microb Cell Factories 15:16. doi:10.1186/s12934-016-0415-9
Bentley R (1990) The shikimate pathway—a metabolic tree with many branches. CRC Crit Rev Biochem Mol Biol 25:307–384. doi:10.3109/10409239009090615
Binder S, Schendzielorz G, Stäbler N, Krumbach K, Hoffmann K, Bott M, Eggeling L (2012) A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol 13:R40. doi:10.1186/gb-2012-13-5-r40
Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R (2005a) Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev 15:125–135. doi:10.1016/j.gde.2005.02.006
Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R (2005b) Transcriptional regulation by the numbers: models. Curr Opin Genet Dev 15:116–124. doi:10.1016/j.gde.2005.02.007
Bjarnason J, Southward CM, Surette MG (2003) Genomic profiling of iron-responsive genes in Salmonella enterica serovar typhimurium by high-throughput screening of a random promoter library. J Bacteriol 185:4973–4982. doi:10.1128/JB.185.16.4973-4982.2003
Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi:10.1126/science.277.5331.1453
Bongaerts J, Krämer M, Müller U, Raeven L, Wubbolts M (2001) Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab Eng 3:289–300. doi:10.1006/mben.2001.0196
Brewster RC, Weinert FM, Garcia HG, Song D, Rydenfelt M, Phillips R (2014) The transcription factor titration effect dictates level of gene expression. Cell 156:1312–1323. doi:10.1016/j.cell.2014.02.022
Carey LB, van Dijk D, Sloot PM, Kaandorp JA, Segal E (2013) Promoter sequence determines the relationship between expression level and noise. PLoS Biol 11:e1001528. doi:10.1371/journal.pbio.1001528
Chen G, Yanofsky C (2003) Tandem transcription and translation regulatory sensing of uncharged tryptophan tRNA. Science 301:211–213. doi:10.1126/science.1084902
Chung CT, Niemela SL, Miller RH (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A 86:2172–2175
Craggs TD (2009) Green fluorescent protein: structure, folding and chromophore maturation. Chem Soc Rev 38:2865–2875. doi:10.1039/b903641p
Cremer J, Eggeling L, Sahm H (1990) Cloning the dapA dapB cluster of the lysine-secreting bacterium Corynebacterium glutamicum. MGG Mol Gen Genet 220:478–480
Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, Keasling JD (2013) Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol 31:1039–1046. doi:10.1038/nbt.2689
Delvigne F, Zune Q, Lara AR, Al-Soud W, Sorensen SJ (2014) Metabolic variability in bioprocessing: implications of microbial phenotypic heterogeneity. Trends Biotechnol 32:608–616. doi:10.1016/j.tibtech.2014.10.002
Eikmanns BJ, Thum-Schmitz N, Eggeling L, Ludtke KU, Sahm H (1994) Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140(Pt 8):1817–1828. doi:10.1099/13500872-140-8-1817
Freed NE, Silander OK, Stecher B, Böhm A, Hardt WD, Ackermann M (2008) A simple screen to identify promoters conferring high levels of phenotypic noise. PLoS Genet 4:e1000307. doi:10.1371/journal.pgen.1000307
Fritz G, Megerle JA, Westermayer SA, Brick D, Heermann R, Jung K, Radler JO, Gerland U (2014) Single cell kinetics of phenotypic switching in the arabinose utilization system of E. coli. PLoS ONE 9:e89532. doi:10.1371/journal.pone.0089532
Gerigk M, Bujnicki R, Ganpo-Nkwenkwa E, Bongaerts J, Sprenger G, Takors R (2002) Process control for enhanced L-phenylalanine production using different recombinant Escherichia coli strains. Biotechnol Bioeng 80:746–754. doi:10.1002/bit.10428
Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi:10.1038/nmeth.1318
Grinter NJ (1998) Developing an L-phenylalanine process. Chem Tech:33–35
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi:10.1016/S0022-2836(83)80284-8
Harper MA, Chen Z, Toy T, Machado IM, Nelson SF, Liao JC, Lee CJ (2011) Phenotype sequencing: identifying the genes that cause a phenotype directly from pooled sequencing of independent mutants. PLoS ONE 6:e16517. doi:10.1371/journal.pone.0016517
Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T (2006) Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2(2006):0007. doi:10.1038/msb4100049
Heatwole VM, Somerville RL (1991) The tryptophan-specific permease gene, mtr, is differentially regulated by the tryptophan and tyrosine repressors in Escherichia coli K-12. J Bacteriol 173:3601–3604
Herrmann KM, Weaver LM (1999) The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 50:473–503. doi:10.1146/annurev.arplant.50.1.473
Hiraga S, Ito K, Matsuyama T, Ozaki H, Yura T (1968) 5-methyltryptophan-resistant mutations linked with the arginine G marker in Escherichia coli. J Bacteriol 96:1880–1881
Hoffmann K, Grünberger A, Lausberg F, Bott M, Eggeling L (2013) Visualization of imbalances in sulfur assimilation and synthesis of sulfur-containing amino acids at the single-cell level. Appl Environ Microbiol 79:6730–6736. doi:10.1128/AEM.01804-13
Hou Y, Hossain GS, Li J, Shin HD, Liu L, Du G (2015) Production of phenylpyruvic acid from L-phenylalanine using an L-amino acid deaminase from Proteus mirabilis: comparison of enzymatic and whole-cell biotransformation approaches. Appl Microbiol Biotechnol 99:8391–8402. doi:10.1007/s00253-015-6757-0
Iizuka R, Yamagishi-Shirasaki M, Funatsu T (2011) Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal Biochem 414:173–178. doi:10.1016/j.ab.2011.03.036
Kasian PA, Pittard J (1984) Construction of a tyrP-lac operon fusion strain and its use in the isolation and analysis of mutants derepressed for tyrP expression. J Bacteriol 160:175–183
Kensy F, Zang E, Faulhammer C, Tan RK, Büchs J (2009) Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb Cell Factories 8:31. doi:10.1186/1475-2859-8-31
Keren L, Zackay O, Lotan-Pompan M, Barenholz U, Dekel E, Sasson V, Aidelberg G, Bren A, Zeevi D, Weinberger A, Alon U, Milo R, Segal E (2013) Promoters maintain their relative activity levels under different growth conditions. Mol Syst Biol 9:701. doi:10.1038/msb.2013.59
Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Schroder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD (2013) EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612. doi:10.1093/nar/gks1027
Lange C, Mustafi N, Frunzke J, Kennerknecht N, Wessel M, Bott M, Wendisch VF (2012) Lrp of Corynebacterium glutamicum controls expression of the brnFE operon encoding the export system for L-methionine and branched-chain amino acids. J Biotechnol 158:231–241. doi:10.1016/j.jbiotec.2011.06.003
Leuchtenberger W, Huthmacher K, Drauz K (2005) Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol 69:1–8. doi:10.1007/s00253-005-0155-y
Liu D, Evans T, Zhang F (2015a) Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng 31:35–43. doi:10.1016/j.ymben.2015.06.008
Liu D, Xiao Y, Evans BS, Zhang F (2015b) Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth Biol 4:132–140. doi:10.1021/sb400158w
Luo Y, Zhang T, Wu H (2014) The transport and mediation mechanisms of the common sugars in Escherichia coli. Biotechnol Adv 32:905–919. doi:10.1016/j.biotechadv.2014.04.009
Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25:1203–1210. doi:10.1093/nar/25.6.1203
Mahr R, Frunzke J (2016) Transcription factor-based biosensors in biotechnology: current state and future prospects. Appl Microbiol Biotechnol 100:79–90. doi:10.1007/s00253-015-7090-3
Mahr R, Gätgens C, Gätgens J, Polen T, Kalinowski J, Frunzke J (2015) Biosensor-driven adaptive laboratory evolution of L-valine production in Corynebacterium glutamicum. Metab Eng 32:184–194. doi:10.1016/j.ymben.2015.09.017
Mäkelä J, Kandhavelu M, Oliveira SM, Chandraseelan JG, Lloyd-Price J, Peltonen J, Yli-Harja O, Ribeiro AS (2013) In vivo single-molecule kinetics of activation and subsequent activity of the arabinose promoter. Nucleic Acids Res 41:6544–6552. doi:10.1093/nar/gkt350
Miller, J. H., 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory.
Mustafi N, Bott M, Frunzke J (2015) Genetically-encoded biosensors for strain development and single cell analysis of Corynebacterium glutamicum. In: Burkovski A (ed) Corynebacterium glutamicum: from systems biology to biotechnological applications. Caister Academic Press, Norfolk, p. 190
Mustafi N, Grünberger A, Kohlheyer D, Bott M, Frunzke J (2012) The development and application of a single-cell biosensor for the detection of L-methionine and branched-chain amino acids. Metab Eng 14:449–457. doi:10.1016/j.ymben.2012.02.002
Mustafi N, Grünberger A, Mahr R, Helfrich S, Noh K, Blombach B, Kohlheyer D, Frunzke J (2014) Application of a genetically encoded biosensor for live cell imaging of L-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS ONE 9:e85731. doi:10.1371/journal.pone.0085731
Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441:840–846. doi:10.1038/nature04785
Ogino T, Garner C, Markley JL, Herrmann KM (1982) Biosynthesis of aromatic compounds: 13C NMR spectroscopy of whole Escherichia coli cells. Proc Natl Acad Sci U S A 79:5828–5832. doi:10.1073/pnas.79.19.5828
Ohlendorf R, Vidavski RR, Eldar A, Moffat K, Möglich A (2012) From dusk till dawn: one-plasmid systems for light-regulated gene expression. J Mol Biol 416:534–542. doi:10.1016/j.jmb.2012.01.001
Payne JW (1977) Transport and hydrolysis of peptides by microorganisms. CIBA Found Symp:305–334
Pittard AJ, Davidson BE (1991) TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol 5:1585–1592
Pittard J (1996) The various strategies within the TyrR regulation of Escherichia coli to modulate gene expression. Genes Cells 1:717–725. doi:10.1111/j.1365-2443.1996.tb00012.x
Pittard J, Camakaris H, Yang J (2005) The TyrR regulon. Mol Microbiol 55:16–26. doi:10.1111/j.1365-2958.2004.04385.x
Polen T, Krämer M, Bongaerts J, Wubbolts M, Wendisch VF (2005) The global gene expression response of Escherichia coli to L-phenylalanine. J Biotechnol 115:221–237. doi:10.1016/j.jbiotec.2004.08.017
Robijns SC, Roberfroid S, Van Puyvelde S, De Pauw B, Uceda Santamaria E, De Weerdt A, De Coster D, Hermans K, De Keersmaecker SC, Vanderleyden J, Steenackers HP (2014) A GFP promoter fusion library for the study of Salmonella biofilm formation and the mode of action of biofilm inhibitors. Biofouling 30:605–625. doi:10.1080/08927014.2014.907401
Rodriguez A, Martinez JA, Flores N, Escalante A, Gosset G, Bolivar F (2014) Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb Cell Factories:13. doi:10.1186/s12934-014-0126-z
Rüffer N, Heidersdorf U, Kretzers I, Sprenger GA, Raeven L, Takors R (2004) Fully integrated L-phenylalanine separation and concentration using reactive-extraction with liquid-liquid centrifuges in a fed-batch process with E. coli. Bioprocess Biosyst Eng 26:239–248. doi:10.1007/s00449-004-0354-4
Sambrook J, MacCallum P, Russell D (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Santos CN, Stephanopoulos G (2008) Melanin-based high-throughput screen for L-tyrosine production in Escherichia coli. Appl Environ Microbiol 74:1190–1197. doi:10.1128/AEM.02448-07
Sariaslani FS (2007) Development of a combined biological and chemical process for production of industrial aromatics from renewable resources. Annu Rev Microbiol 61:51–69. doi:10.1146/annurev.micro.61.080706.093248
Sarsero JP, Pittard AJ (1991) Molecular analysis of the TyrR protein-mediated activation of mtr gene expression in Escherichia coli K-12. J Bacteriol 173:7701–7704
Schallmey M, Frunzke J, Eggeling L, Marienhagen J (2014) Looking for the pick of the bunch: high-throughput screening of producing microorganisms with biosensors. Curr Opin Biotechnol 26:148–154. doi:10.1016/j.copbio.2014.01.005
Schendzielorz G, Dippong M, Grünberger A, Kohlheyer D, Yoshida A, Binder S, Nishiyama C, Nishiyama M, Bott M, Eggeling L (2014) Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth Biol 3:21–29. doi:10.1021/sb400059y
Schneider WP, Ho SK, Christine J, Yao M, Marra A, Hromockyj AE (2002) Virulence gene identification by differential fluorescence induction analysis of Staphylococcus aureus gene expression during infection-simulating culture. Infect Immun 70:1326–1333
Semsey S, Krishna S, Sneppen K, Adhya S (2007) Signal integration in the galactose network of Escherichia coli. Mol Microbiol 65:465–476. doi:10.1111/j.1365-2958.2007.05798.x
Siedler S, Stahlhut SG, Malla S, Maury J, Neves AR (2014) Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab Eng 21:2–8. doi:10.1016/j.ymben.2013.10.011
Silva-Rocha R, de Lorenzo V (2012) Broadening the signal specificity of prokaryotic promoters by modifying cis-regulatory elements associated with a single transcription factor. Mol BioSyst 8:1950–1957. doi:10.1039/c2mb25030f
Simmonds S, Griffith DD (1961) Metabolism of phenylalanine-containing peptide amides in Escherichia coli. J Bacteriol 83:256–263
Sprenger GA (2006) Aromatic amino acids. In: Steinbüchel A (ed) Microbiology monographs. vol. Amino acid biosynthesis—pathways, regulation and metabolic engineering. Springer-Verlag, Berlin Heidelberg
Sprenger GA (2007) From scratch to value: engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl Microbiol Biotechnol 75:739–749. doi:10.1007/s00253-007-0931-y
Tabor, J. J., Groban, E. S., Voigt, C. A., 2009, Performance characteristics for sensors and circuits used to program E. coli. In: Lee, S. Y., (Ed.), Systems biology and biotechnology of Escherichia coli. Springer Science & Business Media B.V.
Uchiyama T, Abe T, Ikemura T, Watanabe K (2005) Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol 23:88–93. doi:10.1038/nbt1048
Uchiyama T, Watanabe K (2008) Substrate-induced gene expression (SIGEX) screening of metagenome libraries. Nat Protoc 3:1202–1212. doi:10.1038/nprot.2008.96
van Summeren-Wesenhagen PV, Marienhagen J (2015) Metabolic engineering of Escherichia coli for the synthesis of the plant polyphenol pinosylvin. Appl Environ Microbiol 81:840–849. doi:10.1128/AEM.02966-14
Vargas-Tah A, Martinez LM, Hernandez-Chavez G, Rocha M, Martinez A, Bolivar F, Gosset G (2015) Production of cinnamic and p-hydroxycinnamic acid from sugar mixtures with engineered Escherichia coli. Microb Cell Factories 14:6. doi:10.1186/s12934-014-0185-1
Vrljic M, Sahm H, Eggeling L (1996) A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826. doi:10.1046/j.1365-2958.1996.01527.x
Weickert MJ, Adhya S (1992) Isorepressor of the gal regulon in Escherichia coli. J Mol Biol 226:69–83. doi:10.1016/0022-2836(92)90125-4
Weiner M, Albermann C, Gottlieb K, Sprenger GA, Weuster-Botz D (2014) Fed-batch production of L-phenylalanine from glycerol and ammonia with recombinant Escherichia coli. Biochem Eng J 83:62–69. doi:10.1016/j.bej.2013.12.001
Whipp MJ, Pittard AJ (1977) Regulation of aromatic amino acid transport systems in Escherichia coli K-12. J Bacteriol 132:453–461
Williams B, Paigen K (1968) Paradoxical effect of weak inducers on the lac operon of Escherichia coli. J Bacteriol 96:1774–1782
Wilson TJ, Argaet VP, Howlett GJ, Davidson BE (1995) Evidence for two aromatic amino acid-binding sites, one ATP-dependent and the other ATP-independent, in the Escherichia coli regulatory protein TyrR. Mol Microbiol 17:483–492
Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci U S A 111:11299–11304. doi:10.1073/pnas.1406401111
Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods 3:623–628. doi:10.1038/nmeth895
Zeevi D, Sharon E, Lotan-Pompan M, Lubling Y, Shipony Z, Raveh-Sadka T, Keren L, Levo M, Weinberger A, Segal E (2011) Compensation for differences in gene copy number among yeast ribosomal proteins is encoded within their promoters. Genome Res 21:2114–2128. doi:10.1101/gr.119669.110
Zhang F, Carothers JM, Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol 30:354–359. doi:10.1038/nbt.2149
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by the German Federal Ministry of Education and Research (BMBF OptoSys grant 031A167B) and the Helmholtz Association (Helmholtz grant VH-NG-716).
Conflict of interest
Julia Frunzke declares that she has no conflict of interest. Regina Mahr declares that she has no conflict of interest. Raphael Freiherr von Boeselager declares that he has no conflict of interest. Johanna Wiechert declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF .98 mb)
Rights and permissions
About this article
Cite this article
Mahr, R., von Boeselager, R.F., Wiechert, J. et al. Screening of an Escherichia coli promoter library for a phenylalanine biosensor. Appl Microbiol Biotechnol 100, 6739–6753 (2016). https://doi.org/10.1007/s00253-016-7575-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7575-8