Abstract
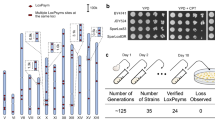
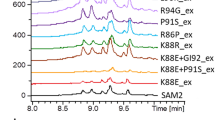
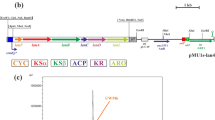
Streptomyces albus J1074 is a derivative of the S. albus G1 strain defective in SalG1 restriction–modification system. Genome sequencing of S. albus J1074 revealed that the size of its chromosome is 6.8 Mb with unusually short terminal arms of only 0.3 and 0.4 Mb. Here we present our attempts to evaluate the dispensability of subtelomeric regions of the S. albus J1074 chromosome. A number of large site-directed genomic deletions led to circularization of the S. albus J1074 chromosome and to the overall genome reduction by 307 kb. Two spontaneous mutants with an activated carotenoid cluster were obtained. Genome sequencing and transcriptome analysis indicated that phenotypes of these mutants resulted from the right terminal 0.42 Mb chromosomal region deletion, followed by the carotenoid cluster amplification. Our results indicate that the right terminal 0.42 Mb fragment is dispensable under laboratory conditions. In contrast, the left terminal arm of the S. albus J1074 chromosome contains essential genes and only 42 kb terminal region is proved to be dispensable. We identified overexpressed carotenoid compounds and determined fitness costs of the large genomic rearrangements.



Similar content being viewed by others
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Baltz RH (2010) Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37:759–772
Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bilyk B, Weber S, Myronovskyi M, Bilyk O, Petzke L, Luzhetskyy A (2013) In vivo random mutagenesis of streptomycetes using mariner-based transposon Himar1. Appl Microbiol Biotechnol 97:351–359
Chater KF, Wilde LC (1980) Streptomyces albus G mutants defective in the SalGI restriction–modification system. J Gen Microbiol 116:323–334
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER (2009) BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6:677–681
Dykhuizen DE (1990) Experimental studies of natural selection in bacteria. Annu Rev Ecol Syst 21:373–398
Fedoryshyn M, Welle E, Bechthold A, Luzhetskyy A (2008) Functional expression of the Cre recombinase in actinomycetes. Appl Microbiol Biotechnol 78:1065–1070
Feng Z, Wang L, Rajski SR, Xu Z, Coeffet-LeGal MF, Shen B (2009) Engineered production of iso-migrastatin in heterologous Streptomyces hosts. Bioorg Med Chem 17:2147–2153
Fischer G, Decaris B, Leblond P (1997) Occurrence of deletions, associated with genetic instability in Streptomyces ambofaciens, is independent of the linearity of the chromosomal DNA. J Bacteriol 179:4553–4558
Fuciman M, Chábera P, Zupcanová A, Hríbek P, Arellano JB, Vácha F, Psencík J, Polívka T (2010) Excited state properties of aryl carotenoids. Phys Chem Chem Phys 12:3112–3120
Gravius B, Bezmalinović T, Hranueli D, Cullum J (1993) Genetic instability and strain degeneration in Streptomyces rimosus. Appl Environ Microbiol 59:2220–2228
Green MR, Sambrook J (2012) Molecular cloning: a laboratory manual, 4th edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Gullón S, Olano C, Abdelfattah MS, Braña AF, Rohr J, Méndez C, Salas JA (2006) Isolation, characterization, and heterologous expression of the biosynthesis gene cluster for the antitumor anthracycline steffimycin. Appl Environ Microbiol 72:4172–4183
Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF (2004) λ Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol 54:107–128
Huang CH, Lin YS, Yang YL, Huang SW, Chen CW (1998) The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol Microbiol 28:905–916
Kieser T, Bibb M, Buttner M, Chater K, Hopwood D (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Kim S-Y, Zhao P, Igarashi M, Sawa R, Tomita T, Nishiyama M, Kuzuyama T (2009) Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afford a diterpene cyclase and two p450 hydroxylases. Chem Biol 16:736–743
Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H (2010) Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A 107:2646–2651
Krügel H, Krubasik P, Weber K, Saluz HP, Sandmann G (1999) Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim Biophys Acta 1439:57–64
Lee HS, Ohnishi Y, Horinouchi S (2001) A σB-like factor responsible for carotenoid biosynthesis in Streptomyces griseus. J Mol Microbiol Biotechnol 3:95–101
Lombó F, Velasco A, Castro A, de la Calle F, Braña AF, Sánchez-Puelles JM, Méndez C, Salas JA (2006) Deciphering the biosynthesis pathway of the antitumor thiocoraline from a marine actinomycete and its expression in two streptomyces species. Chembiochem Eur J Chem Biol 7:366–376
Makitrynskyy R, Rebets Y, Ostash B, Zaburannyi N, Rabyk M, Walker S, Fedorenko V (2010) Genetic factors that influence moenomycin production in streptomycetes. J Ind Microbiol Biotechnol 37:559–566
Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R (2011) AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–346
Moradas-Ferreira P, Costa V, Piper P, Mager W (1996) The molecular defences against reactive oxygen species in yeast. Mol Microbiol 19:651–658
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–185
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A (2011) β-Glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol 77:5370–5383
Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Olson JA, Krinsky NI (1995) Introduction: the colorful, fascinating world of the carotenoids: important physiologic modulators. FASEB J 9:1547–1550
Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A 98:12215–12220
Pósfai G, Plunkett G 3rd, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR (2006) Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046
Qin Z, Cohen SN (1998) Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol Microbiol 28:893–903
Rauland U, Glocker I, Redenbach M, Cullum J (1995) DNA amplifications and deletions in Streptomyces lividans 66 and the loss of one end of the linear chromosome. Mol Gen Genet 246:37–44
Rodriguez L, Oelkers C, Aguirrezabalaga I, Braña AF, Rohr J, Méndez C, Salas JA (2000) Generation of hybrid elloramycin analogs by combinatorial biosynthesis using genes from anthracycline-type and macrolide biosynthetic pathways. J Mol Microbiol Biotechnol 2:271–276
Sandmann G, Kuhn S, Böger P (1998) Evaluation of structurally different carotenoids in Escherichia coli transformants as protectants against UV-B radiation. Appl Environ Microbiol 64:1972–1974
Schumann G, Nürnberger H, Sandmann G, Krügel H (1996) Activation and analysis of cryptic crt genes for carotenoid biosynthesis from Streptomyces griseus. Mol Gen Genet 252:658–666
Takaichi S (2000) Characterization of carotenes in a combination of a C18 HPLC column with isocratic elution and absorption spectra with a photodiode-array detector. Photosynth Res 65:93–99
Takano H, Obitsu S, Beppu T, Ueda K (2005) Light-induced carotenogenesis in Streptomyces coelicolor A3(2): identification of an extracytoplasmic function sigma factor that directs photodependent transcription of the carotenoid biosynthesis gene cluster. J Bacteriol 187:1825–1832
Volff JN, Altenbuchner J (1998) Genetic instability of the Streptomyces chromosome. Mol Microbiol 27:239–246
Wendt-Pienkowski E, Huang Y, Zhang J, Li B, Jiang H, Kwon H, Hutchinson CR, Shen B (2005) Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J Am Chem Soc 127:16442–16452
Widenbrant EM, Tsai H-H, Chen CW, Kao CM (2008) Spontaneous amplification of the actinorhodin gene cluster in Streptomyces coelicolor involving native insertion sequence IS466. J Bacteriol 190:4754–4758
Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, Moore BS (2007) Molecular basis for chloronium-mediated meroterpene cyclization: cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J Biol Chem 282:16362–16368
Yanai K, Murakami T, Bibb M (2006) Amplification of the entire kanamycin biosynthetic gene cluster during empirical strain improvement of Streptomyces kanamyceticus. Proc Natl Acad Sci U S A 103:9661–9666
Zhou M, Jing X, Xie P, Chen W, Wang T, Xia H, Qin Z (2012) Sequential deletion of all the polyketide synthase and nonribosomal peptide synthetase biosynthetic gene clusters and a 900-kb subtelomeric sequence of the linear chromosome of Streptomyces coelicolor. FEMS Microbiol Lett 333:169–179
Acknowledgements
This work was supported through funding from the ERC starting grant EXPLOGEN No. 281623 to AL. The authors thank the Waters company (Milford, USA) for the UPCC measurements of carotenoids.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 520 kb)
Rights and permissions
About this article
Cite this article
Myronovskyi, M., Tokovenko, B., Brötz, E. et al. Genome rearrangements of Streptomyces albus J1074 lead to the carotenoid gene cluster activation. Appl Microbiol Biotechnol 98, 795–806 (2014). https://doi.org/10.1007/s00253-013-5440-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5440-6