Abstract
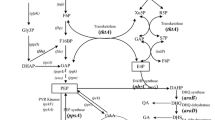
Pseudomonas chlororaphis GP72 is a root-colonizing biocontrol strain isolated from the green pepper rhizosphere that synthesizes two phenazine derivatives: phenazine-1-carboxylic acid (PCA) and 2-hydroxyphenazine (2-OH-PHZ). The 2-OH-PHZ derivative shows somewhat stronger broad-spectrum antifungal activity than PCA, but its conversion mechanism has not yet been clearly revealed. The aim of this study was to clone and analyze the phenazine biosynthesis gene cluster in this newly found strain and to improve the production of 2-OH-PHZ by gene disruption and precursor addition. The conserved phenazine biosynthesis core operon in GP72 was cloned by PCR, and the unknown sequences located upstream and downstream of the core operon were detected by random PCR gene walking. This led to a complete isolation of the phenazine biosynthesis gene cluster phzIRABCDEFG and phzO in GP72. Gene rpeA and phzO were insertionally mutated to construct GP72AN and GP72ON, respectively, and GP72ANON collectively. The inactivation of rpeA resulted in a fivefold increase in the production of PCA, as well as 2-OH-PHZ. The addition of exogenous precursor PCA to the broth culture, to determine the conversion efficiency of PCA to 2-OH-PHZ under current culture conditions, revealed that PCA had a positive feedback effect on its own accumulation, leading to enhanced synthesis of both PCA and 2-OH-PHZ. The production of 2-OH-PHZ by GP72AN increased to about 170 μg ml−1, compared with just 5 μg ml−1 for the wild type. The hypothesis of biosynthetic pathway for 2-OH-PHZ from PCA was confirmed by identification of 2-hydroxyphenazine-1-carboxylic acid as an intermediate in the culture medium of the high-phenazine producing GP72AN mutant.






Similar content being viewed by others
References
Ausubel FM, Kingstons RE, Moore DD, Seidman JG, Smith JA, Struhl K (1995) Short protocols in molecular biology. Wiley, New York
Brodhagen M, Henkels MD, Loper JE (2004) Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 70:1758–1766
Cabeza ML, Aguirre A, Soncini FC, Véscovi EG (2007) Induction of RpoS degradation by the two-component system regulator RstA in Salmonella enteric. J Bacteriol 189:7335–7342
Chancey ST, Wood DW, Pierson EA, Pierson LS III (2002) Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30–84 in the wheat rhizosphere. Appl Environ Microbiol 68:3308–3314
Chin-A-Woeng TFC, Thomas-Oates JE, Lugtenberg BJJ, Bloemberg GV (2001) Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol Plant-Microb Interact 14:1006–1015
Chin-A-Woeng TFC, Bloemberg GV, Lugtenberg BJJ (2003) Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol 157:503–523
Delaney SM, Mavrodi DV, Bonsall RF, Thomashow LS (2001) PhzO, a gene for biosynthesis of 2-hydroxylated phenazine compounds in Pseudomonas aureofaciens 30–84. J Bacteriol 183:318–327
Dmitri VM, Robert FB, Shannon MD, Marilyn JS, Greg P, Thomashow LS (2001) Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183(21):6454–6465
Dwivedi D, Johri BN (2003) Antifungals from fluorescent Pseudomonads: biosynthesis and regulation. Curr Sci 85:1693–1703
Girard G, Rij ETV, Lugtenberg BJJ, Bloemberg GV (2006) Regulatory roles of psrA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology 152:43–58
Hoang TT, Karkhoff-Schweizer RAR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86
Keen NT, Tamaki S, Kobayashi D, Keen DT (1998) Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J Laboratoral Clin Med 44:301–307
Kumaresan K, Subramanian M, Vaithiyanathan S, Sevagaperumal N, GOPAL C, Dilantha FWG (2005) Broad spectrum action of phenazine against active and dormant structures of fungal pathogens and root knot nematode. Arch Phytopathol Plant Prot 38:69–76
Liu HM, Dong D, Peng HS, Zhang XH, Xu XQ (2006a) Genetic diversity of phenazine- and pyoluteorin-producing pseudomonads isolated from green pepper rhizosphere. Arch Microbiol 185:91–98
Liu HM, He YJ, Jiang HX (2006b) Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Curr Microbiol 54:302–306
Liu HM, Yan A, Zhang XH, Xu YQ (2008) Phenazine-1-carboxylic acid biosynthesis in Pseudomonas chlororaphis GP72 is positively regulated by the sigma factor RpoN. World J Microbiol Biotechnol 24:1961–1966
Maddula VSRK, Pierson EA, Pierson LS III (2008) Atering the ratio of phenazines in Pseudomonas chlororaphis strain 30–84: effects on biofilm formation and pathogen inhibition. J Bacteriol 190:2759–2766
Morales VM, Backman A, Bagdasarian M (1991) A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47
Pilhofer M, Bauer AP, Schrallhammer M, Richter L, Ludwig W, Schleifer KH, Petroni G (2007) Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res 35:e135
Price-Whelan A, Dietrich LEP, Newman DK (2006) Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol 2:71–78
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Habor Laboratory, New York
Samina M, Baig DN, Jamil F, Weselowski B, Lazarovits G (2009) Characterization of a phenazine and hexanoyl homoserine lactone producing Pseudomonas aurantiaca strain PB-St2, isolated from sugarcane stem. J Microbiol Biotechnol 19:1688–1694
Schnider-Keel U, Seematter A, Maurhofer M, Blumer C, Duffy B, Gigot-Bonnefoy C, Reimmann C, Notz R, Défago G, Haas D, Keel C (2000) Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol 182:1215–1225
Shen J, Huang L, Peng HS, Hu HB, Zhang XH (2009) The reaction mechanism for phenazine-1-carboxylic acid to 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Abstr J Biosci Bioeng 108:114–134
Smirnov VV, Kiprianova EA (1990) Bacteria of Pseudomonas genus. Naukova Dumka, Kiev, pp 100–111
Whistler CA, Pierson LS III (2003) Repression of phenazine antibiotic production in Pseudomonas aureofaciens strain 30–84 by RpeA. J Bacteriol 185:3718–3725
Veselova MA, Klein Sh, Bass IA, Lipasova VA, Metlitskaya AZ, Ovadis MI, Chernin LS, Khmel IA (2008) Quorum sensing systems of regulation, synthesis of phenazine antibiotics, and antifungal activity in rhizospheric bacterium Pseudomonas chlororaphis 449. Russ J Genet 44:1400–1408
Acknowledgements
This study was supported by the 973 Programs of China (no. 2009CB118906), 863 Programs of China (no. 2006AA10A2009), National Nature Science Foundation of China (NSFC) (nos. 30821005 and 30870075), and Ph.D. Programs Foundation of Ministry of Education of China (no. 20090073110052).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, L., Chen, MM., Wang, W. et al. Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Appl Microbiol Biotechnol 89, 169–177 (2011). https://doi.org/10.1007/s00253-010-2863-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2863-1