Abstract
While cultivation is a convenient way of proliferating and understanding bacteria, studies have shown the formation of nonculturable cells in nonspore-forming bacteria in response to environmental stress and thus in turn have generated immense interest. Whether these cells are in a state of dormancy or in a stage preceding cell death has been considered of paramount importance for the past couple of decades. In this study, osmotic-stress-induced dormant bacterial cells were separated by cell sorting and revived by osmotic down-shift in the absence of nutrients, source(s) that potentially could supply nutrients, and/or the external addition of resuscitation factor(s). Reversal of dormancy followed a definite pattern akin to population asynchrony of dormant cells, and the phenomenon was observed across three species, namely, Enterobacter sp. strain mcp11b, Klebsiella pneumonia strain mcp11d and Escherichia coli. In addition, our study precisely forecasted the presence of multiple subpopulations in dormant cells, which is explained by an emerging theory of survival mechanisms in stressful environments. These observations reveal that the state of dormancy induced by environmental stress in these nonspore-forming bacteria is “reversible” and also implies that it is an orderly and spontaneous adaptation to circumvent adverse conditions.
Similar content being viewed by others
Introduction
Nonspore-forming bacteria, when subjected to stress, lose their culturability leading to a transient dormancy state [described as viable but nonculturable (VBNC), nonculturable (NC), or active but nonculturable (ABNC)]. Conceptual and methodological intricacies of this dormancy state have been extensively debated (see reviews by Roszak and Colwell 1987; Kell et al. 1998; McDougald et al. 1998; Mukamolova et al. 2003; Nystrom 2003 and references cited therein). Despite evidence on the revival of growth in these dormant cells, a number of basic and logical questions have been raised as to whether this nonculturability is a dormancy state of cell adaptation to stress or a stage preceding cell death, partly owing to the unpersuasive revitalization potential. Studies also showed the molecular and functional dissimilarity of stressed (Chan et al. 1998), VBNC cells (Heim et al. 2002) from the NC state induced by “starvation”, which was demonstrated using isolated NC cells (Desnues et al. 2003), leading to the suggestion that NC cells might have been formed as a result of stochastic deterioration. Nonetheless, while co-crystallization of DNA with the stress induced proteins was proposed as a generic defense strategy to environmental stress in Escherichia coli (Wolf et al. 1999), a resuscitation-promoting factor(s) (Rpf) was also identified in long-term stationary phase cultures of Micrococcus luteus (Mukamolova et al. 1998).
It is well known that most of the nonspore-forming bacteria form dormant cells under stress. Combined with a proposition that a cell’s adaptive response is nonspecific for stress factors (see review by Kültz 2005 and references cited therein), they could even be considered as a basic prototype to understand the bacterial remodeling potential to environmental stress. However, this necessitates prior demonstration of these dormant cells as a distinct state of adaptation. In addition, interfering signals from other coexisting physiological types may impede further experimentation and interpretation. To date, there is no evidence on the revival of growth demonstrated on isolated dormant cells, although a number of empirical methods involving, for example, reduced levels of gene expression, morphological changes, and/or other related phenotypes were considered to demonstrate and/or detect these dormant cells in a wide variety of bacterial species (see review by Nystrom 2003 and references cited therein), thus leading to a significant progress in the field. Despite evidence on revival, other co-existing cell populations, detritus, and dissolved nutrients were shown to have potential implications on the revival process of these dormant cells. Hence, in the past, studies proved challenging due to the lack of appropriate tools to differentially study these cells.
The focus of the present study was to understand (1) whether the dormant cells formed in response to osmotic stress in Enterobacter sp. strain mcp11b, Klebsiella pneumonia strain mcp11d, and E. coli can be separated; (2) if separated, can they be revitalized (resuscitated and re-grown); (3) if revitalized, does the phenomenon occur in all three species; and (4) whether the process of revitalization is abrupt, arbitrary, or follows a definite pattern, which would comprehensively reveal whether this unique physiological state in these nonspore-forming bacteria formed in response to osmotic stress is a dormant state of adaptation to circumvent stress.
Materials and methods
Bacterial strains
Bacterial strains include Enterobacter sp. strain mcp11b and K. pneumonia strain mcp11d, which were isolated by screening polluted waters using media for Fecal Coliforms (m-FC) agar (BD, Maryland, USA) void of rosolic acid. Polymerase chain reaction-amplified products of chromosomal DNA of isolates mcp11b and mcp11d were sequenced using the ABI Prism 3100 DNA sequencer and ABI Prism BigDye Terminator cycle sequencing ready-reaction kit. Primers used to amplify 16s rRNA gene include 27F, 530F, 926F, 519R, 907R, and 1492R (Lane 1991). Sequence similarity search was performed using basic local alignment search tool (BLAST; http://www.ncbi.nlm.nih.gov/BLAST/). The 16s rRNA gene sequences of mcp11b and mcp11d were submitted to the GenBank (http://www.ncbi.nlm.nih.gov/), and they were assigned accession numbers of EF419181 and EF419182 and identified as Enterobacter sp. and K. pneumonia. These isolates were also deposited in publicly accessible culture collections namely NCIMB (Aberdeen, Scotland) and NCCB (The Netherlands Culture Collection of Bacteria, Utrecht, Netherlands). They were assigned accession numbers, NCIMB 14479 and NCCB 100231 and NCIMB 14480 and NCCB 100232 for Enterobacter sp. mcp11b and K. pneumonia strain mcp11d, respectively. The strain of E. coli was obtained from American Type Culture Collection (ATCC 700891).
Microcosms and osmotic stress
Waters used in the microcosm experiments were obtained from the Marina Bay in Singapore. Waters were collected in pre-sterilized containers and filtered through membrane filters with pore size of 0.45 μm (Gelman Sciences, Ann Arbor, MI, USA) and sterilized by autoclaving. Pre-washed cells grown in Luria–Bertani (LB) of Enterobacter sp. strain mcp11b, K. pneumonia strain mcp11d, and E. coli were inoculated individually to 0.5-L pre-sterilized bottles containing sterile microcosm waters to a final cell density of around 108 cells mL−1 to induce saline stress in static conditions at 30°C. The salinity of the waters used in the microcosms was 30 ppt. The optimal salinity that is isotonic to nonhalophilic bacteria is around 8 ppt (salinity of a standard physiological saline). Therefore, nonhalophilic bacterium, namely, Enterobacter sp., K. pneumonia, and E. coli, upon introduction in to natural saline waters at 30 ppt, will experience 3.75 times higher salinity resulting in osmotic stress. We then used the flow cytometric procedure (Sachidanandham et al. 2005) to measure rigid viable cells (RVC) as an indicator of the presence of dormant cells.
Flow cytometric analysis and fluorescence-activated cell sorting
To determine the viability of the culture, cells were stained using the LIVE/DEAD BacLight kit (Molecular Probes, Eugene/Portland, OR, USA) using the procedure described previously (Sachidanandham et al. 2005). In this previous study, using E. coli and an environmental isolate (H03N1), we had shown that LIVE/DEAD BacLight kit can differentiate and precisely enumerate live cells in the liquid nitrogen treatment processes in hyper-saline microcosms. Several studies have successfully used LIVE/DEAD BacLight kit for the measurement of bacterial viability in environmental isolates (e.g., Haznedaroglua et al. 2008; Portaels et al. 2008). These observations, together with our earlier work conducted on osmotically stressed cultures of E. coli and coliform environmental isolate (H03N1) (Sachidanandham et al. 2005), forecasted that no abnormalities in the staining of LIVE/DEAD BacLight kit in the viability measurements were observed (also see Fig. 5, live and dead cell count of Enterobacter sp. strain mcp11b assayed by BacLight). This kit was also used to estimate viable cell count of bacteria directly on various types of environmental samples successfully (see review by Trevors 2003 and references therein). Abnormalities are usually associated with inappropriate use of reagents. It should be noted that freshly reconstituted reagents need to be used. Storing of reconstituted reagents and reusing them will provide inaccurate enumeration and distribution patterns.
LIVE/DEAD BacLight reagents were reconstituted in physiological saline solution “as and when needed”. Flow cytometric analysis and fluorescence-activated cell sorting were carried out using a FACSCalibur (Becton Dickinson, San Jose, CA, USA), which is equipped with four-color multi-parameter analytical and data acquisition capabilities. It is equipped with a 15-mW argon ion laser, which produces blue light of 488 nm. The following optical settings were used: side scatter (SS), 488 nm/10 band pass (bp); forward scatter (FS), 488 nm/10 bp; FL1, 530 nm/30 bp (green fluorescence); and FL3, 650 nm long pass (red fluorescence). The primary and secondary thresholds were set on the FL1 and SS, respectively. The argon ion laser can excite propidium iodide (PI) and SYTO 9 in the analysis of BacLight-stained cells. Cells are hydrodynamically focused in the flow cytometer (FCM) to a single file flow through, followed by laser excitation at 488 nm. Within the flow cell of the FCM, a relatively slow-moving sample stream is injected into a rapidly moving sheath (microcosm water) stream. Sheath is a term used to describe a liquid that is used to align the cells of interest in a single-file flow through so that they can be interrogated with appropriate laser to study the cell properties (Shapiro 2003). The sample stream is drawn rapidly from the injection point into a narrow fast-moving stream within the sheath stream. Controlling the velocity of these streams, the cells within the center stream can be aligned to single file and interrogated by the laser. The emitted fluorescence is captured by photomultiplier tubes (PMT).
LIVE/DEAD BacLight kit differentiates cells with intact cell membranes stained by SYTO 9 (i.e., scored as live) from those with damaged membranes stained by both SYTO 9 and PI (i.e., scored as dead). PI quenches the SYTO 9 fluorescence in dead cells. Apart from parameters such as SS and FS, the green emission with respect to SYTO 9 binding and red emission with respect to PI binding were captured in FL1 and FL3 PMTs, respectively. The instrument design of FACSCalibur ensures that sorting occurs in a completely enclosed, aerosol-free environment for enhanced bio-safety, which has been extensively used for fluorescence-activated cell sorting (FACS) in the past. To achieve optimal data rates (150 to 500 cells/s) for cell sorting, samples were diluted using microcosm waters. Sorting was always performed in “single-cell” sort mode using the same water as sheath used in the preparation of microcosm cultures. “Gating” is a procedure in which specific cell populations can be enumerated in the flow cytometric analysis and/or separated in the FACS procedures. Target live-cell populations were gated in the software for sorting. Fluorescence microspheres of 2 μm in size (Polyscience, USA) were used both as size and volumetric calibration standards. Data acquisition was carried out using the software CELLQuest (Becton Dickinson), and the data analysis was performed in WinMDI-2.8 (http://facs.scripps.edu/software.html). Total viable cell count (TVC) was assayed by directly staining the microcosm cultures using LIVE/DEAD kit and RVC (cells that retain viability after liquid nitrogen treatment) were assayed after selective inactivation of culturable cells (CC; Sachidanandham et al. 2005). Bacterial cells stained with BacLight were spotted onto a slide and examined using an epifluorescence microscope with thousand times magnification (BX40, Olympus, Japan). Cells were viewed under blue light excitation (488 nm) using the U-25ND6 and U-25ND25 filters (Olympus, Japan).
Revitalization of dormant cells
Six hundred to 650 ml of sorted cells in the sheath was aliquoted to equal volumes in sterile tubes and centrifuged at 3,250 × g (4,000 rpm) for 15 min. Baseline revitalization and/or sort “contaminants of culturable cells” (contaminant in the sort process refers to culturable cells of the same strain), if any, were studied by instantaneously plating an equal volumetric fraction of sorted cells on LB agar using the sterile sheath water as diluent. Incubation up to 24 h at 37°C did not show any visible colonies in all three strains; hence, the incubation time was prolonged up to 48 h, and the colonies were counted. Further incubation until the completion of the entire revitalization assay for a given sorting did not show any variation in the count. Colonies in these plates, if any, could be a result of contaminating culturable cells (CCSC) in the sorting or weak resuscitation in the sheath, as the sorting process usually takes 3 to 4 h. Thus, these were taken as CCSC. In parallel, a portion of the samples collected from the microcosm cultures for sorting was also plated (before liquid nitrogen treatment) using the identical procedure as that of the assay of sort contaminants, and this resulted in profuse colonies of CC. Single-cell sort mode sorts exclusively target cells within the sort loop. If a target cell appears together with a nontarget cell within the sort loop, it will be rejected, thus leading to high sort purity. An aliquot of sorted cells was re-stained using BacLight and examined under microscope for the live cells. Cells were sorted from arbitrarily chosen time periods of 8, 9, or 12 days of Enterobacter sp. strain mcp11b, K. pneumonia strain mcp11d, and E. coli microcosm cultures respectively or specified therein. Sorted cells resuspended in phosphate-buffered saline (PBS; salinity, 8 ppt) were incubated in a shaking bath at 100 rpm at 37°C. Aliquots were drawn periodically, serially diluted using sterile PBS, and plated on LB agar. Counting was performed after 48 h incubation at 37°C in all the analyses, and the value of CCSC, if any, was subtracted from these values resulting in revitalized cell counts (RVC). The relative ratio of culturable cells to that of resuscitated cells were estimated using the formula:
where, CCSC = CC in the sorted cells, RVc = highest number of revitalized cells (cell count after PBS resuscitation, CCSC).
Results
Flow cytometric measurement and fluorescence-activated cell sorting of dormant cells
Bacterial species chosen for the present study include Enterobacter sp. strain mcp11b, K. pneumonia strain mcp11d, and E. coli. While all the three species belong to the coliform group, it is to be noted that Enterobacter sp. strain mcp11b and K. pneumonia strain mcp11d are environmental isolates and E. coli is a laboratory strain. Osmotic stress was introduced in natural saline waters. FACS, together with fluorescence probes, has been used as a method of cell separation (see review by Davey and Kell 1996 and references cited therein) for further analysis. Our flow cytometric method of detection of dormant cells were carried out as described in “Materials and methods.” The subset of populations emitting green fluorescence (live cells) (Figs. 1b, 2b, and 3b) was gated in the software, resulting in the selective elimination of nontarget cells (dead cells) in the FACS cell sorting.
Flow cytometric analysis, FACS cell separation and revitalization of Enterobacter sp. strain mcp11b. a Flow cytometric profile of distribution of various cell types, namely P1 (dead), P2 (live), and P3 (live) in microcosm cultures. b Flow cytometric profile of sample after subjected to selective inactivation. c Levels of total viable cells (TVC), culturable cells (CC), and rigid viable cells (RVC) (cells used for separation by FACS). d Revitalization of FACS separated dormant cells as a function of time
Flow cytometric analysis, FACS cell separation and revitalization of K. pneumonia strain mcp11d. a Flow cytometric profile of distribution of various cell types, namely P1 (dead) and P3 (live) in microcosm cultures. b Flow cytometric profile of sample after subjected to selective inactivation. c Levels of total viable cells (TVC), culturable cells (CC), and rigid viable cells (RVC) (cells used for separation by FACS). d Revitalization of FACS separated dormant cells as a function of time
Flow cytometric analysis, FACS cell separation and revitalization of E. coli. a Flow cytometric profile of distribution of various cell types, namely P1 (dead) and P3 (live) in microcosm cultures. b Flow cytometric profile of sample after subjected to selective inactivation. c Levels of total viable cells (TVC), culturable cells (CC), and rigid viable cells (RVC) (cells used for separation by FACS). d Revitalization of FACS separated dormant cells as a function of time
Choice of sheath
Since our procedures involve studies concerning osmotically sensitive cells, selection of appropriate sheath is important. It should be isotonic to ensure that no or minimal osmotic disturbances occur during sorting and cell concentration steps. Therefore, the natural saline water used for the preparation of microcosm cultures was used as sheath in the FACS procedures.
Selective staining of various cell types by BacLight
Samples were drawn from the saline-stressed cultures as described in the “Materials and methods” and stained using BacLight and analyzed in the FCM to examine the presence of various cell types. As evident from the flow cytometric dot plots of Enterobacter sp. (Fig. 1a), K. pneumonia (Fig. 2a), and E. coli (Fig. 3a), various cell types, namely, dead and live cells, were resolved based on the respective fluorescence emission. Cell populations designated as P1 banded at the red PMT (red fluorescence) as a result of quenching of SYTO 9 fluorescence by PI; hence, they are scored as “dead cells.” Cell populations that were banded at the green PMT (green fluorescence) imply that they were selectively stained by SYTO 9 and impermeable to PI in the BacLight assay. Therefore, they were scored as “live.”
Population asynchrony of dormant cells
In the BacLight-based standard viability assays, we encounter one single population for each of the category of live and dead cells banding at the green and red PMTs, respectively (data not shown). In this study involving osmotically stressed cells, we found that there are multiple subpopulations within a population of Enterobacter sp. (see P3 in Fig. 1a), K. pneumonia (Fig. 2a), and E. coli (Fig. 3a). It is evident that these subpopulations band as “small clusters” and are diagonally arrayed parallel to the x-axis (green fluorescence). Each small cluster represents a subpopulation within a single physiological state of live cells. It is interesting to note that these multiple subpopulations are “conserved” in the RVC of Enterobacter sp., (Fig. 1b), K. pneumonia (Fig. 2b), and E. coli (Fig. 3b). This phenomenon is designated as “population asynchrony.”
Microdiversity of dormant cells based on DNA levels
SYTO 9 of BacLight is a DNA-specific fluorochrome. The uptake of syto9 is linearly related to the levels of intracellular DNA in bacterial cells, which is generally reflected in the intensity of green fluorescence measured by the green PMT in a FCM. The FCM dot plots are divided into four decades in logarithmic scale. “Decade” is a term used in flow cytometry to indicate a division in the dot plot (shown as thin lines, for instance in Fig. 1a). In general, SYTO 9-stained cells banding at the first decade will have the lowest level of DNA when compared to cells in the fourth decade. Therefore, cell populations appearing within P3 vary in their overall intracellular DNA levels, as reflected by their stretched distribution in the third and fourth decades of green fluorescence shown in the flow cytometric dot plots of Enterobacter sp. (Fig. 1a), K. pneumonia (Fig. 2a), and E. coli (Fig. 3a). This is designated as “microdiversity.”
Rigid viable cells
To find out the relative levels of TVC, RVC, and CC, enumeration was carried out using the respective procedures described in the “Materials and methods,” and the results are shown in Figs. 1c, 2c, and 3c for Enterobacter sp., K. pneumonia, and E. coli, respectively. In our previous study, we had shown that rigid viable cells (described as ABNC cells) formed in response to osmotic stress are direct indicators of the presence of dormant cells, and they acquired mechanical resistance to overcome osmotic stress (Sachidanandham et al. 2005). Therefore, samples containing RVC were further subjected to FACS to separate the dormant cells, if any, and assess whether they were reversible.
Gating and cell separation
A schematic representation of the whole approach of FACS is shown in Fig. 4. It is evident from the flow cytometric dot plots that the population designated as P3 is sharply differentiated from other cell types (P1 and/or P2) in Enterobacter sp. (Fig. 1b), K. pneumonia (Fig. 2b), and E. coli (Fig. 3b). P1 banded at the red-PMT predominantly in the first and second decades. P3 banded in the third and/or fourth decades in the green-PMT. These populations largely differed in their relative fluorescence with respect to green (x-axis) and red PMTs (y-axis). This made it convenient to draw this population (P3) out of other interfering cell types in the FACS. It is to be noted that if this cell type is merged with other cell types (we have not encountered this in our measurements so far), the method of FACS will be unsuitable. Therefore, “gating” of clearly separated cell types in the dot plots is a key step in the successful FACS target-specific cells separation process. FACSCalibur is equipped with “self containment” for sorting biosafety level 2 organisms (human pathogens), where sorting occurs in a completely enclosed aerosol-free environment. During the sorting process, a significant amount of liquid is pumped together with the sorted dormant cells. Therefore, centrifugation was used to further concentrate these dormant cells.
Schematic diagram of FACS cell separation and revitalization of dormant cells. [i] Coexistence of culturable cells (CC) and nonculturable dormant cells in microcosm cultures. [ii] After selective inactivation by rapid freeze/thaw, cell separation by FACS. [iii] Resuscitation by osmotic downshift in PBS, and [iv] re-growth on solid medium. The osmotic down-shift involved reduction in salinity by 3.75 times
Choice and validation of revitalization media
Subsequent to the FACS separation, the resuscitations need to be performed in an exclusive medium that should offer favorable conditions for resuscitation without allowing cell multiplication. The medium should offer optimal osmolarity and be void of direct and indirect sources of nutrients. If the elevated salinity and resulting osmotic stress (30 ppt) is the environmental factor that had driven these nonspore-forming bacteria to form dormant cells, providing an optimal salinity to the FACS separated dormant cells of these nonhalophilic bacteria may possibly revive growth and/or multiplication potential. Therefore, PBS was chosen, a frequently used medium in laboratories, to maintain bacterial cells. PBS contains 3.75 times lower salinity (8 ppt) than the microcosm water. Note that we have not tested/used any “resuscitation factor(s)” in our revitalization experiments.
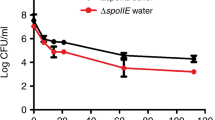
Resuscitation is a frequently used terminology to describe the reversal of dormant cells in conventional single-step procedures. We use the term revitalization to describe the two-step process that includes resuscitation in PBS, followed by re-growth in plate cultures. Despite the fact that there is no basis of bacterial cell multiplication in PBS, it was validated with exponentially grown Enterobacter sp. strain mcp11b (Fig. 5). Reversal of dormancy, if any, was carried out in the validated PBS.
FACS separated cells were revitalized by instantaneously resuspending in PBS. Aliquots of samples were periodically drawn during the resuscitation process in PBS and plated onto agar plates to enumerate revitalized cells, if any. The enumerated cell counts of samples collected at various time intervals from the revitalization media were plotted. It was observed that the revitalized cell count increased as a function of time in Enterobacter sp. (Fig. 1d), K. pneumonia (Fig. 2d), and E. coli (Fig. 3d). It is also obvious from these data that the profiles of revitalization are “sigmoidal” for all three bacterial species. The dormant cells were isolated from the population P3, which exhibited traits such as “population asynchrony” and “microdiversity.” Therefore, we suggest that the sigmoid profile of revitalization may possibly be attributed to these two traits of the dormant cells. As the process of revitalization is invariably observed in all three species used in this study, the phenomenon can be considered nonspecies specific.
Recovery levels of dormant cells
The relative ratios of culturable cells to that of revitalized cells in the FACS sorting and revitalization were computed using the formula shown in the method and found to be 99.47%, 99.54%, and 99.3% for Enterobacter sp., K. pneumonia, and E. coli, respectively. As per this formula, >99% of the FACS sorted live cells exist in dormant form, which are not readily culturable before the resuscitation in PBS. It also implies that 0.53%, 0.46%, and 0.7% of FACS-sorted cells of Enterobacter sp., K. pneumonia, and E. coli, respectively, were sort contaminants of culturable cells. It is to be noted that we could not achieve a batch of FACS-sorted cells totally void of culturable cells even in FACS runs with minimum data rate and gating cells at the right end of FL1. The FACS method described in this paper was aimed at pure forms of dormant cells. Therefore, “single cell” sort mode of the FACSCalibur was used. Sorting in this mode follows a “probability” that even a target cell, if appears together with a nontarget cell within the “sort loop,” will be rejected resulting in the highest purity. Therefore, sort stoichiometry and the relative levels of dormant cells with respect to the overall population cannot be established using this method. It should be noted that the populations described in the formula shown in the “Materials and methods” uses the number of FACS sorted cells cultured on plates before PBS resuscitation (CCSC) and after PBS resuscitation (RVc).
These experimental results imply that hyper-salinity-imparted osmotic stress is the primary environmental stress that can drive nonhalophilic bacterial species, namely, Enterobacter sp. strain mcp11b, K. pneumonia strain mcp11d, and E. coli, to form dormant cells. Using the method of FACS, these dormant cells can be readily separated. Furthermore, they are revitalized by osmotic downshift resulting in cell division/cell multiplication as implied by growth on plate cultures. Therefore, dormancy state acquired by these nonspore-forming bacteria is an adaptation to circumvent environmental assault by osmotic stress.
Discussion
Revitalization—choice of PBS and plate cultures
Studies have shown that in low-temperature-stressed microcosms, temperature upshift can allow nonculturable cells to resuscitate (e.g., Du et al. 2007). On the other hand, some of the studies have also shown that it could possibly be due to bacteria growth (Coutard et al. 2007). With the help of resuscitation, it was also hypothetically shown that VBNC cells can support the rest of the surviving cells (Arana et al. 2007), while they lack intrinsic regeneration potential. These studies, together with many earlier work, forecasted two notions on the nonculturable cells, which include that (1) they can regenerate growing cells as opposed to (2) they lack intrinsic regeneration potential. Hence, this has strongly warranted in our study to decouple growth from resuscitation in process-specific environments using robust techniques. Growth is driven by bacterial-multiplication-utilizing substrate. On the other hand, resuscitation is a transformation process with which cells transform from one state (nonculturable) to another (culturable). The experimental strategies and methodologies used in our study take advantage of flow cytometry and FACS cell separation techniques. Some of the methodological steps need to be optimally designed to avoid experimental ambiguity. For instance, nutritional interference in the revitalization process can allow coexisting culturable cells to grow. We have observed that 0.53%, 0.46%, and 0.7% of FACS-sorted cells of Enterobacter sp., K. pneumonia, and E. coli, respectively, were sort contaminants of culturable cells. While it is evident that PBS is void of any direct or indirect sources of nutrients and no external addition of resuscitation factor was made in our study, we have further validated PBS using Enterobacter sp. (Fig. 5). It is also evident from the validation experiment that there is no cell growth occurring in this medium, hence, providing a nutrient-free environment (void of direct or indirect source of nutrients) for the process of resuscitation. Therefore, revitalization was undertaken in a process specific environment.
Choosing the method to evaluate the revitalized cells is another important step in a successful revitalization experiment. There are innumerable methods available to measure “live” bacterial cells. Most of them can measure small levels of “nonviable cells” as well (Sheridan et al. 1998). Therefore, “culturing” remains appropriate for assessing revitalized live cells originating from dormancy. Recent molecular evidence points to the fact that the environment on an agar plate is primitive and stressful (Cuny et al. 2007). Hence, dormant cells might decide that the prevailing conditions on an agar surface is unfavorable for growth, while revitalized cells (Figs. 1d, 2d, and 3d) expected to have similar physiological state as that of exponentially growing cells, readily form colonies. Our study implied that 99.47%, 99.54%, and 99.3% (computed using the formula shown in the “Materials and methods”) of FACS-separated rigid viable cells of Enterobacter sp., K. pneumonia, and E. coli, respectively, were transformed into reversible dormancy state. Therefore, the phenomenon of formation and revitalization was found to be nonspecies specific.
Some of the earlier studies conducted using in situ resuscitation method (Coutard et al. 2007 and references cited therein) had shown that dead cells can promote the growth of the residual cells giving an appearance of resuscitation. While the methodology used in our study largely differed from these studies, implications of dead cells on the bacterial growth process of coexisting viable cells cannot be neglected. This was investigated in our study in a medium void of substrate for bacterial growth, namely PBS (see “Materials and methods”). Growth in a culture system can occur only when the growth rate is much greater than death rate (Bailey and Ollis 1986; Pirt 1975). The resuscitation medium used in our study, namely PBS, was further investigated by introducing live bacterial cells of Enterobacter sp. mcp11b and progressively following the viable and dead cell dynamics. In principal, upon introducing bacterial cells into a medium: (1) if growth rate is greater than death rate, the bacterial culture should grow resulting in the increase in the initial cell count. A medium exhibiting this profile is unsuitable for resuscitation, as it can select growth over resuscitation. (2) If growth rate is equal to death rate, the viable cell count should remain constant. While there could be no net growth in this medium, it can still allow cells to grow to compensate the loss of viable cells, hence unsuitable for resuscitation. (3) If growth rate much lower or zero when compared to death rate, this medium can never support bacterial growth. Therefore, a medium with this quality would be highly suitable for resuscitation. It is evident that the process observed in PBS shown in Fig. 5 is explained by category 3. Therefore, this medium cannot support bacterial growth.
A proposition also existed that dead cells can allow live cells to grow. If we closely examine our data (Fig. 5), the levels of dead cells were found to be 6.26 × 106 at the beginning of the cultivation, which had risen to 2.87 × 107 at the end. On the other hand, the levels of live cells were 6.21 × 107 at the beginning, which had decreased to 1.32 × 107 (see also Fig. 5). In other words, during the entire incubation periods, both viable and dead cells co-existed in the same medium; however, no bacterial growth was observed. In fact, the viable cell count constantly decreased during the progression of incubation. Therefore, our data obtained in PBS (which was used as a resuscitation medium in our study) dismissed the earlier proposition that dead cells can promote growth of coexisting live cells. Our results further confirmed that earlier observations of dead cell and detritus interference were confined to the respective method of in situ resuscitation and cannot be generalized.
Another proposition was that detritus can allow coexisting live cells to grow, giving an appearance of resuscitation. This can be examined by analyzing the stoichiometry of our data (Fig. 5). It was interesting to note that there was a decrease in the total cell count from 6.84 × 107 to 4.18 × 107. This further implied that approximately 39% of cells introduced into the system had lysed. In spite of massive detritus and lysate that should have been released due the cell lysis, there was a decrease in the viable cell count (Fig. 5). This proved beyond doubt that detritus did not promote cell growth in the medium used for resuscitation. Therefore, our data dismissed the interference of dead cells and detritus in the subsequent resuscitation process in PBS. Hence, earlier observations on the dead cells and detritus interference (see review by Nystrom 2003 and references therein) cannot be extrapolated and generalized.
There is immense interest in these dormant cells during the past couple of decades. A basic outline can be drawn on certain properties of these dormant cells (McDougald et al. 1998; Mukamolova et al. 2003; Roth et al. 1998), which include (1) viable nongrowing cells can form in response to environmental stress in nonspore-forming bacteria, (2) revival of culturability ought to be shown in the absence of nutrients, source(s) that can potentially supply nutrients and/or external addition of resuscitation factor(s), and (3) cells should revitalize in response to reduced stress level(s) (reversal of adverse environmental conditions). The revitalization of Enterobacter sp. strain mcp11b, K. pneumonia, and E. coli shown in this study is consistent with these propositions.
Bioprocess implications
It has been shown in many previous studies that addition of nutrients and/or temperature upshift (shown in Vibrio sp.) to revive nonculturable cells in microcosms resulted in the re-growth of few residual culturable cells (see Nystrom 2003 and references therein). These observations are supported by the Monod model, which is one of the most frequently used mathematical models in bioprocesses to describe growth (Bailey and Ollis 1986; Pirt 1975). Media that were used in the past resuscitation studies exhibited S > 0 (see also Eq. 1). Therefore, residual culturable cells multiply rapidly using the residual substrate (S) present in these microcosms or artificial cultures giving a false appearance of resuscitation (Nystrom 2003 and references therein; Supplementary material Fig. S1). Hence, appropriate selection or design of resuscitation medium is important to avoid re-growth of culturable cells.
According to the Monod model,
Where μ = specific growth rate (h−1), S = substrate concentration (g l−1), and K s = substrate saturation constant (g l−1).
This model implies that in order to obtain a sigmoid pattern in bacterial growth, culture should meet two basic prerequisites which include (1) specific growth rate (μ) of a given bacterium should be positive in a medium and (2) substrate concentration (S) should be >0. In contrary, in order for a medium to support exclusive resuscitation (void of growth), (1) a bacterium should exhibit zero or negative growth rate in a given resuscitation medium (implying that the resuscitation medium does not support growth) and (2) substrate concentration should be zero (medium contains no basic resource to support growth).
It is evident that the medium used for resuscitation in our study, namely PBS, contains no substrate for bacterial growth.
Furthermore, sigmoid pattern of bacterial growth is described by:
Where x = cell density (g l−1) and t = time (h).
It is evident from Fig. 5 that specific growth rate in PBS is zero or negative, hence dismissing the possibility of re-growth. Therefore, the sigmoid pattern exhibited by Enterobacter sp. (Fig. 1d), K. pneumonia (Fig. 2d), and E. coli (Fig. 3d) were due a nongrowth phenomenon, which is revitalization (resuscitation in PBS and re-growth in LB agar). These data also imply that nonculturable cells need to be separated from the original medium to show exclusive resuscitation. Furthermore, evaluation of Monod parameter(s) would be helpful in providing key information on the intrinsic capability of a resuscitation medium to support bacterial growth.
It cannot be neglected that what has been shown as revitalization (resuscitation and re-growth) in Enterobacter sp. strain mcp11b (Fig. 1d), K. pneumonia strain mcp11d (Fig. 2d), and E. coli (Fig. 3d) were simply substrate depleted cells (nutrient limitation as per Monod kinetics). These cells had taken some time to grow on LB plates, and they had simply experienced a pronounced time lag. Profound analyses of the regeneration stoichiometry of all three strains (Figs. 1d, 2d, and 3d) provided an answer as to whether they were substrate-depleted cells or survived in dormancy state.
Substrate-depleted cells should respond by growth when supplied with substrate. Similarly, adapted cells for hyper-osmotic conditions should respond when provided with optimal osmotic medium. (1) For instance, the time course revitalization data exhibited a “straight line as function of time”; this would directly imply that all sorted cells responded readily to substrate in LB, rather than osmotic down-shift in PBS. In contrast, what has been observed were sigmoid patterns that were dependent on the time duration of the incubations of cells in PBS rather than growth on LB. Hence, the decisive factor for regeneration was osmotic-down shift rather than substrate.
(2) The incubation of the first aliquot of the plated sample(s) (count shown at 0 h in Figs. 1d, 2d, and 3d) was prolonged throughout the revitalization period (i.e., ≈150 h in Enterobacter sp. mcp11b; ≈80 h in K. pneumonia strain mcp11d and ≈100 h in E. coli). Note that beyond 48 h of incubation there was no further increase in CFU observed in these plates (see also “Materials and methods” for further details). If they were substrate depleted, there should have been a progressive increase in the CFU with time of the 0 h aliquots. Furthermore, the values of culturable cells before passage through PBS (CSsc) was only <0.7%, which was found to be negligible when compared to the levels of regenerated cells after passage through PBS (osmotic downshift >99%) directly disproving the argument that these cells were substrate depleted. Hence, these data proved that the revitalization of cells (Figs. 1d, 2d, and 3d) was in response to osmotic downshift, substantiating further that these cells survived in a dormancy state.
Implications of cellular DNA levels
Bacterial cells can be studied on the basis of intracellular DNA content in flow cytometry with the appropriate use of DNA-specific fluorochromes (Robertson and Button 1989; Shapiro 2003). Our study uses two DNA-based fluorochromes, namely SYTO 9 and PI, in all the flow cytometric analysis and FACS cell separations. Flow cytometric profiles of osmotically stressed dormant cells in three species of nonhalophilic bacteria, namely Enterobacter sp. (Fig. 1a,b), K. pneumonia (Fig. 2a,b), and E. coli (Fig. 3a,b), revealed the following:
-
1.
The distribution of these dormant cells was found to be “diagonally stretched” parallel to the x-axis (green fluorescence) in all three species, i.e., Enterobacter sp., K. pneumonia, and E. coli. It is evident that in the four-decade log scale of the flow cytometric dot plots, cells banding at the “third decade” contain relatively low levels of DNA when compared to the cells at the “fourth decade” within a single population designated as P3 in these figures (except K. pneumonia where two prominent subpopulations banded within the third decade). This implies that there are variations in the intracellular DNA content within these cells, where cells in the third decade contain relatively low levels of DNA when compared to the cells at the fourth decade.
These observations conceptually corroborate earlier reports that co-crystallization of stress-induced proteins with DNA as one of the principle adaptation mechanisms in bacteria to protect genetic material from environmental assault (Wolf et al. 1999). Subsequently, it was also shown that when cells are subjected to stress, DNA modifications are driven by “structural phase transitions,” which require low energy resources (Frenkiel-Krispin et al. 2001). These studies, together with many other works published recently (Ohniwa et al. 2006), forecasted the modification/condensation in DNA as an adaptation mechanism in culturable forms of bacteria. The multiparametric flow cytometric analysis shown in this study shows the spontaneous variations in the intracellular DNA levels in dormant cells resulting in “microdiversity” (a cell population with varying relative levels of DNA content) as a consequence of environmental assault by osmotic stress.
-
2.
Further examination of the flow cytometric profile of the dormant cells of Enterobacter sp. (Fig. 1a,b), K. pneumonia (Fig. 2a,b), and E. coli (Fig. 3a,b) revealed that they are bundled into small subpopulations (small clusters) diagonally arrayed parallel to the x-axis in the logarithmic scale of green fluorescence (see population designated as P3). Each small cluster placed within the third and fourth decades of the flow cytometric dot plots differ from each other in their intrinsic DNA levels. These subpopulations are prominently seen in Enterobacter sp. (see also additional data in Supplementary material Fig. S2 and descriptions therein). An emerging theory on environmental adaptation of bacteria could possibly explain this phenomenon, i.e., in order to occupy different niches of an ecosystem, bacteria create two or more subpopulations in response to environmental stress by “wiring” of a genetic system (see review by Smits et al. 2006 and references cited therein), which was demonstrated in readily culturable bacteria. The total outcome of the adaptation resulted in these subpopulations, which is cellular level evidence that forecasts the possible rigorous phenotypic acquisition routes before multiple phenotypic states. This is in contrast to sporulation in Bacillus, which is a total commitment (i.e., once cells initiate sporulation, the process has to be taken to completion) leading to a homogenous population of spores (Parker et al. 1996). Therefore, the data on population asynchrony demonstrated by flow cytometry, together with many other work forecasting population heterogeneity (Aertsen and Michiels 2005; Booth 2002) support the earlier notion (Frenkiel-Krispin et al. 2001) that environmental defense in nonspore-forming bacteria might leave the cells noncommittal (possibly in multiple phenotypic states). However, it should be noted that we have observed this phenomenon in dormant cells, while the other authors (Aertsen and Michiels 2005; Booth 2002; Frenkiel-Krispin et al. 2001) showed a similar event(s) in culturable forms of bacteria. There is a strong need for bacteria to occupy various ecological niches for nutritional scavenging to prolong their survival (Aertsen and Michiels 2005; Booth 2002; Frenkiel-Krispin et al. 2001). Hence, these heterogenous populations provide survival advantage under stressful environments.
There is a cell-cycle-based notion on the overall heterogeneity (asynchrony and subpopulations) observed in the stressed cultures (Fig. 1a,b). According to this notion, these subpopulations could represent different states of bacteria in the cell cycle and each population in the subpopulations could represent varying levels of chromosome equivalents present in these cells. Nonetheless, several reports in the past had already shown that stressed cells were nongrowing [i.e., upon introducing bacterial cells into the stressor (e.g., hyper-osmotic, low-temperature), no cell growth was observed] (e.g., Arana et al. 2007; Du et al. 2007; see also review by Nystrom 2003 and references cited therein). Furthermore, in all three strains, in samples used for FACS sorting, >70% of the cells were surviving in nonculturable state (see the difference in TVC and CC in Figs. 1c, 2c, and 3c) whose flow cytometric distribution pattern were shown in Figs. 1a, 2a, and 3a. This was largely due to (1) void of growth supporting substrate in the microcosm medium and/or (2) suboptimal physical condition(s) (i.e., 30 ppt salinity) for these nonhalophilic strains. In nongrowing or nonproliferating cultures, the need for the synthesis of new chromosomes remains bleak, and the cells need not undergo cell cycle, as they were largely nonproliferating. It was further shown that these cells cannot proliferate in hyper-osmotic medium, even in the presence of substrates, unless supplied with osmoprotectants (Wood 2006). Subsequent studies also showed that at least in E. coli, ProP protein (osmosensory transporter) sense increasing osmolality and responds by mediating the cytoplasmic accumulation of organic osmolytes (MacMillan et al. 1999; Racher et al. 1999; Romantsov et al. 2007).
Bacterial cells, upon introducing into a hyper-osmotic medium, respond by actively adjusting the distributions of selected solutes across the cytoplamic membrane (Wood 1999). By virtue of this selection process, due to larger molecular weight, perhaps, medium ingredients (sugars and peptides) might remain nonselective in the adapted cells, resulting in nonculturability as evidenced by significant difference observed in the viable cell count (TVC) and culturable cell count (CC) in all three strains (Figs. 1c, 2c, and 3c) in the osmotically stressed cultures of Enterobacter sp. strain mcp11b, K. pneumonia strain mcp11d, and E. coli. This was even true with the sorted cells that they exhibited only <0.7% culturable cells. These evidences prove that it is unlikely that these subpopulations emerged out of formation of chromosome equivalents in nonproliferating cultures.
Recent molecular evidences shed further light on the stress-dependent heterogeneity (Figs. 1a,b, 2a,b, and 3a,b). Even in genetically uniform population of cells, the process that regulate cell function (for example transcription and translation) can, at any time, be differentially activated in different bacterial cells. This was largely shown to be possible by intrinsic and extrinsic variability in gene expression, resulting in heterogeneity that is reversible (see review by Avery 2006 and references therein). It was further shown that larger contribution to the heterogeneity in populations under stress relied on the extrinsic variability in the gene expression (Elowitz et al. 2002; Colman-Lerner et al. 2005). This proposition was also substantiated at translation level (Rosenfeld et al. 2005). Heterogeneity largely helps bacteria to exploit new niches offering distinct survival advantages (Booth 2002; Sumner and Avery 2002). Further evidences also suggested that, in nonspore-forming bacteria, phenotypic heterogeneity is not an irreversible commitment to the new state, so allowing rapid reversion to the original phenotype if appropriate conditions are restored (Thattai and van-Oudenaarden 2004; Kussell and Leibler 2005). This has strongly corroborated our data on stress-dependent nonculturability (see the difference in cell count between TVC and CC in Figs. 1c, 2c, and 3c) revitalization (Figs. 1d, 2d, and 3d) and population asynchrony (Figs. 1a, 2a, and 3a). Emerging evidences also suggested that there is evolutionary selection for diversity-generating mechanisms (Kussell and Leibler 2005; True and Lindquist 2000; Fraser et al. 2004). Studies further implied that degree of heterogeneity or diversity as result of mutagenesis and/or genetic rearrangement is related to the diversity of selective pressure and further forecasted their role in the adaptive evolution (Bjedov et al. 2003). Dynamic rearrangements in the outer membrane architecture, as implied by the proteomic analysis during the formation of VBNC cells of E. coli (Muela et al. 2008), together with versatile and subtle differences on the over all proteome in Enterococcus faecalis (Heim et al. 2002), further strengthens that cells might be undergoing significant subcellular modifications of varying degree resulting in heterogeneity. Hence, a plethora of molecular evidences conceptually corroborated the heterogeneity (asynchrony and subpopulations) observed in the osmotically stressed cultures of Enterobacter sp. strain mcp11b, K. pneumonia strain mcp11d and E. coli, suggesting that heterogeneity possibly offer survival advantage to these strains.
Prototype of stress response
Essentially, flow cytometric analysis of these dormant cells in this study revealed two key modifications at cellular level: (1) microdiversity of cells with varying DNA content and (2) formation of subpopulations within a single major population of cells. These evidences imply that dormant cells had gone through definite molecular remodeling steps before the acquisition of dormancy, and they exist in multiple phenotypic states. Therefore, we suggest that on par with the current study systems involving use of ageing colonies (Bjedov et al. 2003) and long-term stationary cultures (Finkel 2006), this physiological state of dormancy driven by environmental assault by osmotic stress will be a prototype for studies on stress tolerance mechanisms and alternate pathways in nonspore-forming bacteria.
Intracellular DNA modifications (Sachidanandham et al. 2005; Wolf et al. 1999), as implied by the DNA-based banding pattern of dormant cells in the flow cytometric analysis, is an adaptation in nonspore-forming bacteria. A study conducted on the stress response (Frenkiel-Krispin et al. 2001) of E. coli further revealed that the “intracellular DNA sequestration” in stressed cells is neither driven by enzymatic catalysis nor by de novo protein synthesis; hence, these cells might possibly be resistant to conventional antibiotics. Consequently, these dormant cells may fill an important niche in the rationalized understanding of resistance mechanisms in clinical bacteria and profound studies on environmental adaptation, evolution, and emergence of pathogens.
Mechanism(s) of revitalization
Osmosensing in bacteria is conventionally explained by a rapid signal transmission in response to osmotic stress (in milliseconds) driven by mechanosensitive channels (Stokes et al. 2003). If this is the sole mechanism that governs revitalization, the process should occur rapidly. However, we observed that the revitalizations were completed in durations of 80–150 h (Figs. 1d, 2d, and 3d). In an earlier study, it was shown that there were modifications in the peptidoglycan of the bacterial cell wall of these dormant cells (Signoretto et al. 2003). Subsequently, it was suggested that peptidoglycon hydrolysis driven by the antidormancy factor (also called as resuscitation promoting factor) is a key in vivo step in the revitalization of dormant cells, resulting in cell division (Mukamolova et al. 1998; review by Keep et al. 2006 and references therein). Further studies had shown that Rpf-interacting protein was shown to exhibit peptodiglycon hydrolase activity and reactivate dormant cells (Hett et al. 2007). However, these molecular remodeling steps, namely (1) acquiring mechanical resistance by cell wall modification and (2) revitalization by peptidoglycon hydrolysis, are yet to be demonstrated in a single bacterium and with respect to osmotic stress. While the involvement of these mechanisms in the revitalization of osmotically stressed dormant cells cannot be neglected, currently, we can explain the time-dependent reversal of dormancy shown by the “sigmoid” pattern (Figs. 1d, 2d, and 3d) using “population asynchrony” of these cells. We suggest that the population possibly exists in multiple phenotypic states with varying degree of acquired phenotypes. Therefore, the reversal process is time-dependent, resulting in a sigmoid pattern. Further study on the nature of phenotypic acquisition will provide insight on the population distribution at various phenotypic states. Many authors describe this population asynchrony, observed even in culturable forms, using the term “intra-population heterogeneity” (Aertsen and Michiels 2005; Booth 2002). It is proposed that “population asynchrony” improves organismal fitness of bacteria in stressful environments.
In the resuscitation kinetics on the Rpf (also described as autocrine growth factor), it was forecasted that the metabolic activity of the starter (the preculture) to secrete a threshold level of Rpf was necessary to promote growth, implicating that cell–cell communications and/or sensing the threshold levels of Rpf were necessary under certain conditions (Mukamolova et al. 1999). This conceptually explains the longer initial resuscitation lag (≥20 h; Figs. 1d, 2d, and 3d). In fact, the lag was more pronounced in Enterobacter sp. strain mcp11b and K. pneumonia strain mcp11d when compared to the nonpathogenic E. coli. The probable events that could have occurred during the resuscitation lag include regaining homogeneity and triggering the synthesis of metabolites necessary for the resuscitation. At the end of the lag in each of the strains, the threshold concentrations might have been achieved to facilitate the reversal of nonculturability.
The method of cell separation by FACS, apart from demonstrating the revitalization ability of dormancy state at cellular level, paves the way for further experimentation with no or very little interfering signals from other coexisting physiological types (P1) (Fig. 1a,b), which was previously hampering efforts in this area of study.
In this study, for the first time, we have shown that (1) formation (at 30 ppt salinity) and revitalization (8 ppt in PBS) of dormant cells are driven by the magnitude (3.75 times) of osmotic stress in three species of nonhalophilic bacteria, namely, Enterobacter sp., K. pneumonia, and E. coli; (2) the process of reversal of dormancy is demonstrated on sorted nongrowing cells in the absence of nutrients, source(s) that could potentially supply nutrients and/or external addition of resuscitation factor(s); (3) the phenomenon of formation and revitalization of dormant cells are nonspecies specific among the species we have tested; (4) the dormant cell population is “asynchronous” and “diversified” with varying levels of DNA; and (5) the transformation process of reversing the dormancy follows a sigmoid pattern akin to the “population asynchrony”.
These observations lead us to conclude that the state of dormancy demonstrated in these nonspore-forming bacteria in response to osmotic stress is “reversible,” and the process is an orderly and spontaneous adaptation to circumvent environmental stress. In addition, the method of FACS paves the way for further experimentation with no or very little interfering signals from other coexisting physiological types, which was previously hampering efforts in this area of study.
References
Aertsen A, Michiels CW (2005) Diversify or die: generation of diversity in response to stress. Crit Rev Microbiol 31(2):69–78
Arana I, Orruño M, Pérez-Pascual D, Seco C, Muela A, Barcina I (2007) Inability of Escherichia coli to resuscitate from the viable but nonculturable state. FEMS Microbiol Ecol 62(1):1–11
Avery SV (2006) Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4:577–587
Bailey JE, Ollis DF (1986) Biochemical engineering fundamentals, 2nd edn. McGraw-Hill, New York
Bjedov I, Tenaillon O, Gérard B, Souza V, Denamur E, Radman M, Taddei F, Matic I (2003) Stress-induced mutagenesis in bacteria. Science 300(5624):1404–1409
Booth IR (2002) Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int J Food Microbiol 78(1–2):19–30
Chan PF, Foster SJ, Ingham E, Clements MO (1998) The Staphylococcus aureus alternative sigma factor sigmaB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol 180(23):6082–6089
Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R (2005) Regulated cell-to-cell variation in a cell-fate decision system. Nature 437:699–706
Coutard F, Crassous P, Droguet M, Gobin E, Colwell RR, Pommepuy M, Hervio-Heath D (2007) Recovery in culture of viable but nonculturable Vibrio parahaemolyticus: regrowth or resuscitation? ISME J 1:111–120
Cuny C, Maïalène L, Dukan S (2007) Induction of a global stress response during the first step of Escherichia coli plate growth. Appl Environ Microbiol 73(3):885–889
Davey HM, Kell DB (1996) Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev 60(4):641–696
Desnues B, Cuny C, Grégori G, Dukan S, Aguilaniu H, Nyström T (2003) Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep 4(4):400–404
Du M, Chen J, Zhang X, Li A, Li Y (2007) Characterization and resuscitation of viable but nonculturable Vibrio alginolyticus VIB283. Arch Microbiol 188(3):283–288
Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297:1183–1186
Finkel SE (2006) Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 4(2):113–120
Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB (2004) Noise minimization in eukaryotic gene expression. PLoS Biol 2:834–838
Frenkiel-Krispin D, Levin-Zaidman S, Shimoni E, Wolf SG, Wachtel EJ, Arad T, Finkel SF, Kolter R, Minsky A (2001) Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J 20:1184–1191
Haznedaroglua BZ, Bolsterb CH, Walkera SL (2008) The role of starvation on Escherichia coli adhesion and transport in saturated porous media. Water Res 42(6–7):1547–1554
Heim S, Lleo MDM, Bonato B, Guzman CA, Canepari P (2002) The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J Bacteriol 184(23):6739–6745
Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ (2007) A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Molecular Microbiol 66(3):658–668
Keep NH, Wardb JM, Cohen-Gonsaudc M, Henderson B (2006) Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol 14(6):271–276
Kell DB, Kaprelyants AS, Weichart DH, Harwood CR, Barer MR (1998) Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie van Leeuwenhoek 73(2):169–187
Kültz D (2005) Molecular and evolutionary basis of the cellular stress response. Ann Rev Physiol 67:225–257
Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075–2078
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, UK, pp 115–175
MacMillan SV, Alexander DA, Culham DE, Kunte HJ, Marshall EV, Rochon D, Wood JM (1999) The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim Biophys Acta 1420:30–44
McDougald D, Rice SA, Weichart D, Kjelleberg S (1998) Nonculturability: adaptation or debilitation? FEMS Microbiol Ecol 25(1):1–9
Muela A, Seco C, Camafeita E, Arana I, Orrun M, Lopez JA, Barcina I (2008) Changes in Escherichia coli outermembrane subproteome under environmental conditions inducing the viable but nonculturable state. FEMS Microbiol Ecol 64:28–36
Mukamolova GV, Kaprelyants AS, Young DI, Young M, Kell DB (1998) A bacterial cytokine. Proc Natl Acad Sci USA 95(15):8916–8921
Mukamolova GV, Kormer SS, Kell DB, Kaprelyants AS (1999) Stimulation of the multiplication of Micrococcus luteus by an autocrine growth factor. Arch Microbiol 172:9–14
Mukamolova GV, Kaprelyants AS, Kell DB, Young M (2003) Adoption of the transiently non-culturable state—a bacterial survival strategy? Adv Microb Physiol 47:65–129
Nystrom T (2003) Nonculturable bacteria: programmed survival forms or cells at death’s door? Bioessays 25(3):204–211
Ohniwa RL, Morikawa K, Kim J, Ohta T, Ishihama A, Wada C, Takeyasu K (2006) Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J 25(23):5591–5602
Parker GF, Daniel RA, Errington J (1996) Timing and genetic regulation of commitment to sporulation in Bacillus subtilis. Microbiology 142:3445–3452
Pirt SJ (1975) Principles of microbe and cell cultivation. Blackwell, Oxford
Portaels F, Meyers WM, Ablordey A, Castro AG, Chemlal K et al (2008) First cultivation and characterization of Mycobacterium ulcerans from the environment. PLoS Neg Trop Dis 2(3):e178
Racher KI, Voegele RT, Marshall EV, Culham DE, Wood JM, Jung H et al (1999) Purification and reconstitution of an osmosensor: transporter ProP of Escherichia coli senses and responds to osmotic shifts. Biochemistry 38:1676–1684
Robertson BR, Button DK (1989) Characterizing aquatic bacteria according to population, cell size, and apparent DNA content by flow cytometry. Cytometry 10:70–76
Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM (2007) Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbial 64(6):1455–1465
Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB (2005) Gene regulation at the single-cell level. Science 307:1962–1965
Roszak DB, Colwell RR (1987) Survival strategies of bacteria in the natural environment. Microbiol Rev 51(3):365–379
Roth WG, Leckie MP, Dietzler DN (1998) Restoration of colony-forming activity in osmotically stressed Escherichia coli by betaine. Appl Environ Microbiol 54(12):3142–3146
Sachidanandham R, Gin KYH, Poh CL (2005) Monitoring of active but non-culturable bacterial cells by flow cytometry. Biotechnol Bioeng 89(1):24–31
Shapiro HM (2003) Practical flow cytometry, 4th edn. Wiley, Hoboken, NJ
Sheridan GEC, Masters CI, Shallcross JA, Mackey BM (1998) Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol 64(4):1313–1318
Signoretto C, Lleò MM, Tafi CM, Canepari P (2003) Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl Environ Microbiol 66(5):1953–1959
Smits WK, Kuipers OP, Veening J (2006) Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4:259–271
Stokes NR, Murray HD, Subramaniam C, Gourse RL, Louis P, Bartlett W, Miller S, Booth IR (2003) A role for mechanosensitive channels in survival of stationary phase: regulation of channel expression by RpoS. Proc Natl Acad Sci USA 100(26):15959–15964
Sumner ER, Avery SV (2002) Phenotypic heterogeneity: differential stress resistance among individual cells of the yeast Saccharomyces cerevisiae. Microbiology 148:345–351
Thattai M, van-Oudenaarden A (2004) Stochastic gene expression in fluctuating environments. Genetics 167:523–530
Trevors JT (2003) Fluorescent probes for bacterial cytoplasmic membrane research. J Biochem Biophys Methods 57:87–103
True HL, Lindquist SLA (2000) Yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407:477–483
Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A (1999) DNA protection by stress-induced biocrystailization. Nature 400:83–85
Wood JM (1999) Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev 63:230–262
Wood JM (2006) Osmosensing by bacteria. Sci STKE 357:pe43
Acknowledgment
We thank Mr. Yee Woon Kang and Ms. Goh Shin Giek for their assistance in environmental sampling. Funding support by the Ministry of Education, Singapore is acknowledged. We thank all the anonymous reviewers for their valuable suggestions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
As reported, the method of resuscitation involving addition of nutrients to the microcosm cultures or resuscitation in situ, results in the multiplication of residual culturable cells (CC). Therefore, the process of revitalization comprising of resuscitation and re-growth, needs to be carried out in two distinct steps under a “process specific environment” (PPT 196 KB)
Supplementary Fig. S2
P3 cell populations banded as “tiny bean like structures” and diagonally arrayed parallel to the x-axis (green fluorescence). Each tiny bean represents a subpopulation within a single physiological state of live cells. Enterobacter sp.,strain mcp11b after 15 (I) and 22 days (II) incubation in microcosm waters. After selective inactivation, the rigid viable cells with conserved subpopulaions in 15 (III) and 22 (IV) days samples (PPT 278 KB)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sachidanandham, R., Yew-Hoong Gin, K. A dormancy state in nonspore-forming bacteria. Appl Microbiol Biotechnol 81, 927–941 (2009). https://doi.org/10.1007/s00253-008-1712-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1712-y