Abstract
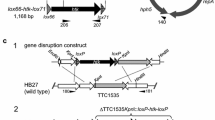
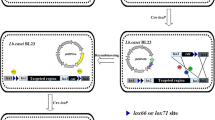
Genome rearrangement is an increasingly important technique to facilitate the understanding of genome functions. A Cre/loxP-mediated deletion system for large-scale genome rearrangements in Corynebacterium glutamicum was developed. By comparative analysis of C. glutamicum R and C. glutamicum 13032 genomes, distinct 14.5-kb and 56-kb regions not essential for cell survival were identified and targeted for deletion. By homologous recombination, loxP sites were integrated at each end of the target region. Deletions between the two chromosomal loxP sites in the presence of Cre recombinase were highly efficient. Accurate deletion was observed in all 96 Cre-expressing strains tested. These deletions represent the largest genomic excisions in C. glutamicum reported to date. Despite the loss of 11 and 58 predicted ORF(s), respectively, upon the deletion of the14.5-kb and 56-kb regions, the cells still exhibited normal growth under standard laboratory conditions. Based on the precision of its deletion, the Cre/loxP system provides a new, efficient genome rearrangement technique for studying C. glutamicum.





Similar content being viewed by others
References
Araki K, Araki M, Yamamura K (1997) Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res 25:868–872
Campo N, Daveran-Mingot ML, Leenhouts K, Ritzenthaler P, Le Bourgeois P (2002) Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis. Appl Environ Microbiol 68:2359–2367
Gopaul DN, Guo F, Duyne GDV (1998) Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J 17:4175–4187
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62:99–109
Itaya M, Tanaka T (1997) Experimental surgery to create subgenomes of Bacillus subtilis 168. Proc Natl Acad Sci 94:5378–5382
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Kramer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Puhler A, Rey DA, Ruckert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25
Kolisnychenko V, Plunkett G IIIrd, Herring CD, Feher T, Posfai J, Blattner FR, Posfai G (2002) Engineering a reduced Escherichia coli genome. Genome Res 12:640–647
Kinoshita S (1985) Glutamic acid bacteria. In: Demain AL, Solomon NA (eds) Biology of industrial microorganisms. Cummings, London, pp 115–146
Kotrba P, Inui M, Yukawa H (2003) A single V317A or V317M substitution in Enzyme II of a newly identified β-glucosidase phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiol 149:1569–1580
Kuhn R, Torres RM (2002) Cre/loxP recombination system and gene targeting. Methods Mol Biol 180:175–204
Kurusu, Y, Kainuma M, Inui M, Satoh Y, Yukawa H (1990) Electroporation-transformation system for Coryneform bacteria by auxotrophic complementation. Aglic Biol Chem 54:443–447
Liebl W (1991) The genus Corynebacterium-nonmedical. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, II. Springer-Verlag, Berlin, Heidelberg, New York, pp 1157–1171
Malumbres M, Mateos LM, Martin JF (1995) Microorganisms for amino acid production: Escherichia coli and corynebacteria. In: Hui YH, Kachatourians GG (eds) Food biotechnology microorganisms, vol 2. VCH, New York, pp 423–469
Murphy E, Huwyler L, do Carmo de Freire Bastos M (1985) Transposon Tn554 complete nucleotide sequence and isolation of transposition-defective and antibiotic sensitive mutants. EMBO J 4:3357–3365
Muyrers JP, Zhang Y, Testa G, Stewart AF (1999) Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res 27:1555–1557
Nakata K, Inui M, Kos PB, Vertès AA, Yukawa H (2003) Vector for the genetic engineering of Corynebacteria. In: Saha BC (ed) Fermentation biotechnology, ACS symposium series, vol 862. American Chemical Society, Washington DC, pp 175–191
Nishio Y, Nakamura Y, Kawarabayasi Y, Usuda Y, Kimura E, Sugimoto S, Matsui K, Yamagishi A, Kikuchi H, Ikeo K, Gojobori T (2003) Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res 13:1572–1579
Ohnishi J, Hayashi M, Mitsuhashi S, Ikeda M (2003) Efficient 40 degrees C fermentation of l-lysine by a new Corynebacterium glutamicum mutant developed by genome breeding. Appl Microbiol Biotechnol 62:69–75
Posfai G, Kolisnychenko V, Bereczki Z, Blattner FR (1999) Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res 27:4409–4415
Qin M, Bayley C, Stockton T, Ow DW (1994) Cre Recombinase-mediated site-specific recombination between plant chromosomes. Proc Natl Acad Sci 91:1706–1710
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning, a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Terasawa M, Inui M, Goto M, Shikata K, Imanari M, Yukawa H (1990) Living cell reaction process for l-isoleucine and l-valine production. J Ind Microbiol 5:289–294
Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1993) Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res Microbiol 144:181–185
Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1994a) Isolation and characterization of IS31831, a transposable element from Corynebacterium glutamicum. Mol Microbiol 11:739–746
Vertes AA, Asai Y, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1994b) Transposon mutagenesis of coryneform bacteria. Mol Gen Genet 245:397–405
Yamagata H, Terasawa M, Yukawa H (1994) A novel industrial process for l-aspartic acid production using an ultrafiltration-membrane. Catalysis Today 22:621–627
Yu BJ, Sung BH, Koob MD, Lee CH, Lee JH, Lee WS, Kim MS, Kim SC (2002) Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat Biotechnol 20:1018–1023
Acknowledgments
We wish to thank Dr Ken-ichi Yamamura for plasmid pBS185. This study was carried out as a part of The Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers by Ministry of Economy, Trade & Industry (METI), and entrusted by New Energy and Industrial Technology Development Organization (NEDO).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Suzuki, N., Tsuge, Y., Inui, M. et al. Cre/loxP-mediated deletion system for large genome rearrangements in Corynebacterium glutamicum. Appl Microbiol Biotechnol 67, 225–233 (2005). https://doi.org/10.1007/s00253-004-1772-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-004-1772-6