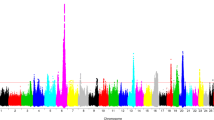
Abstract
A decrease in the incidence of bovine mastitis, the costliest disease in the dairy industry, can be facilitated through genetic marker-assisted selective breeding programs. Identification of genomic variants associated with mastitis resistance is an ongoing endeavor for which genome-wide association studies (GWAS) using high-density arrays provide a valuable tool. We identified single nucleotide polymorphisms (SNPs) in Holstein dairy cattle associated with mastitis resistance in a GWAS by using a high-density SNP array. Mastitis-resistant (15) and mastitis-susceptible (28) phenotypic extremes were identified from 224 lactating dairy cows on commercial dairy farm located in Utah based on multiple criteria of mastitis resistance over an 8-month period. Twenty-seven quantitative trait loci (QTLs) for mastitis resistance were identified based on 117 SNPs suggestive of genome-wide significance for mastitis resistance (p ≤ 1 × 10−4), including 10 novel QTLs. Seventeen QTLs overlapped previously reported QTLs of traits relevant to mastitis, including four QTLs for teat length. One QTL includes the RAS guanyl-releasing protein 1 gene (RASGRP1), a candidate gene for mastitis resistance. This GWAS identifies 117 candidate SNPs and 27 QTLs for mastitis resistance using a selective genotyping approach, including 10 novel QTLs. Based on overlap with previously identified QTLs, teat length appears to be an important trait in mastitis resistance. RASGRP1, overlapped by one QTL, is a candidate gene for mastitis resistance.

Similar content being viewed by others
References
Ashwell MS, Heyen DW, Weller JI, Ron M, Sonstegard TS, van Tassell CP, Lewin HA (2005) Detection of quantitative trait loci influencing conformation traits and calving ease in Holstein-Friesian cattle. J Dairy Sci 88:4111–4119. https://doi.org/10.3168/jds.S0022-0302(05)73095-2
Bonnefont CMD, Toufeer M, Caubet C, Foulon E, Tasca C, Aurel MR, Bergonier D, Boullier S, Robert-Granié C, Foucras G, Rupp R (2011) Transcriptomic analysis of milk somatic cells in mastitis resistant and susceptible sheep upon challenge with Staphylococcus epidermidis and Staphylococcus aureus. BMC Genomics 12:208. https://doi.org/10.1186/1471-2164-12-208
Brand B, Hartmann A, Repsilber D, Griesbeck-Zilch B, Wellnitz O, Kühn C, Ponsuksili S, Meyer HHD, Schwerin M (2011) Comparative expression profiling of E. coli and S. aureus inoculated primary mammary gland cells sampled from cows with different genetic predispositions for somatic cell score. Genet Sel Evol 43:24. https://doi.org/10.1186/1297-9686-43-24
Buitenhuis B, Poulsen NA, Gebreyesus G, Larsen LB (2016) Estimation of genetic parameters and detection of chromosomal regions affecting the major milk proteins and their post translational modifications in Danish Holstein and Danish Jersey cattle. BMC Genet 17:114. https://doi.org/10.1186/s12863-016-0421-2
Bush WS, Moore JH (2012) Chapter 11: genome-wide association studies. https://doi.org/10.1371/journal.pcbi.1002822
Clark SA, van der Werf J (2013) Genomic best linear unbiased prediction (gBLUP) for the estimation of genomic breeding values. pp 321–330
Darvasi A (1997) The effect of selective genotyping on QTL mapping accuracy. Mamm Genome 8:67–68. https://doi.org/10.1007/s003359900353
Detilleux JC (2002) Genetic factors affecting susceptibility of dairy cows to udder pathogens. Vet Immunol Immunopathol 88:103–110. https://doi.org/10.1016/S0165-2427(02)00138-1
Dohoo IR, Smith J, Andersen S, Kelton DF, Godden S, Mastitis Research Workers’ Conference (2011) Diagnosing intramammary infections: evaluation of definitions based on a single milk sample. J Dairy Sci 94:250–261. https://doi.org/10.3168/jds.2010-3559
Gao X, Becker LC, Becker DM, Starmer JD, Province MA (2010) Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol 34:100–105. https://doi.org/10.1002/gepi.20430
Guey LT, Kravic J, Melander O, Burtt NP, Laramie JM, Lyssenko V, Jonsson A, Lindholm E, Tuomi T, Isomaa B, Nilsson P, Almgren P, Kathiresan S, Groop L, Seymour AB, Altshuler D, Voight BF (2011) Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genet Epidemiol Swedish Res Counc (Scania Diabetes Regist) 35:236–246. https://doi.org/10.1002/gepi.20572
Halasa T, Huijps K, Østerås O, Hogeveen H (2007) Economic effects of bovine mastitis and mastitis management: a review. Vet Q 29:18–31. https://doi.org/10.1080/01652176.2007.9695224
Harmon RJ, Anderson KL, Kindahl H et al (1994) Physiology of mastitis and factors affecting somatic cell counts. J Dairy Sci 77:2103–2112. https://doi.org/10.3168/jds.S0022-0302(94)77153-8
Holmbeg M, Andersson-Eklund L (2004) Quantitative trait loci affecting health traits in Swedish dairy cattle. J Dairy Sci 87:2653–2659. https://doi.org/10.3168/jds.S0022-0302(04)73391-3
Hu ZL, Park CA, Wu XL, Reecy JM (2013) Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res 41:D871–D879. https://doi.org/10.1093/nar/gks1150
Kaneene JB, Scott Hurd H (1990) The national animal health monitoring system in Michigan. III. Cost estimates of selected dairy cattle diseases. Prev Vet Med 8:127–140. https://doi.org/10.1016/0167-5877(90)90006-4
Kühn C, Reinhardt F, Schwerin M (2008) Marker assisted selection of heifers improved milk somatic cell count compared to selection on conventional pedigree breeding values. Arch Tierz Dummerstorf 51:23–32
National Mastitis Council (1999) Laboratory Handbook on Bovine Mastitis, Revised. National Mastitis Council, Madison, WI
Lander ES, Botstein S (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–674. https://doi.org/10.1038/hdy.2014.4
Meredith BK, Berry DP, Kearney F, Finlay EK, Fahey AG, Bradley DG, Lynn DJ (2013) A genome-wide association study for somatic cell score using the Illumina high-density bovine beadchip identifies several novel QTL potentially related to mastitis susceptibility. Front Genet 4:229. https://doi.org/10.3389/fgene.2013.00229
Nash DL, Rogers GW, Cooper JB, Hargrove GL, Keown JF, Hansen LB (2000) Heritability of clinical mastitis incidence and relationships with sire transmitting abilities for somatic cell score, udder type traits, productive life, and protein yield. J Dairy Sci 83:2350–2360. https://doi.org/10.3168/jds.S0022-0302(00)75123-X
Odegård J, Klemetsdal G, Heringstad B et al (2003) Genetic improvement of mastitis resistance: validation of somatic cell score and clinical mastitis as selection criteria. J Dairy Sci 86:4129–4136. https://doi.org/10.3168/jds.S0022-0302(03)74027-2
Pearson TA, Manolio TA (2008) How to interpret a genome-wide association study. JAMA Stat Res Methods; Genet Genet 29929911:1335–1344. https://doi.org/10.1001/jama.299.11.1335
Philipsson J, Ral G, Berglund B (1995) Somatic cell count as a selection criterion for mastitis resistance in dairy cattle. Livest Prod Sci 41:195–200. https://doi.org/10.1016/0301-6226(94)00067-H
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
R Core Team RF for SC (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/
Reneau JK, Andrews RJ, Kitchen BJ et al (1986) Effective use of dairy herd improvement somatic cell counts in mastitis control. J Dairy Sci 69:1708–1720. https://doi.org/10.3168/jds.S0022-0302(86)80590-2
Reyher KK, Dohoo IR (2011) Diagnosing intramammary infections: evaluation of composite milk samples to detect intramammary infections. J Dairy Sci 94:3387–3396. https://doi.org/10.3168/jds.2010-3907
Rupp R, Boichard D (2003) Genetics of resistance to mastitis in dairy cattle. 671. Vet Res 34:671–688. https://doi.org/10.1051/vetres:2003020
Sahana G, Guldbrandtsen B, Thomsen B, Lund MS (2013) Confirmation and fine-mapping of clinical mastitis and somatic cell score QTL in Nordic Holstein cattle. Anim Genet 44:620–626. https://doi.org/10.1111/age.12053
Santman-Berends IM, Olde Riekerink RG, Sampimon OC et al (2012) Incidence of subclinical mastitis in Dutch dairy heifers in the first 100 days in lactation and associated risk factors. J Dairy Sci 95:2476–2484. https://doi.org/10.3168/jds.2011-4766
Schnabel RD, Sonstegard TS, Taylor JF, Ashwell MS (2005) Whole-genome scan to detect QTL for milk production, conformation, fertility and functional traits in two US Holstein families. Anim Genet 36:408–416. https://doi.org/10.1111/j.1365-2052.2005.01337.x
Schultz LH (1977) Somatic cells in milk-physiological aspects and relationship to amount and composition of Milk. J of’Food Prot 40:125–131
Sears PM, Smith BS, English PB, Herer PS, Gonzalez RN (1990) Shedding pattern of Staphylococcus aureus from bovine intramammary infections. J Dairy Sci 73:2785–2789. https://doi.org/10.3168/jds.S0022-0302(90)78964-3
Seegers H, Fourichon C, Beaudeau F (2003) Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res 34:475–491
Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M (2012) An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet 44:825–830. https://doi.org/10.1038/ng.2314
Seykora AJ, McDaniel BT (1986) Genetics statistics and relationships of teat and udder traits, somatic cell counts, and milk production. J Dairy Sci 69:2395–2407. https://doi.org/10.3168/jds.S0022-0302(86)80679-8
Sodeland M, Kent MP, Olsen HG, Opsal MA, Svendsen M, Sehested E, Hayes BJ, Lien S (2011) Quantitative trait loci for clinical mastitis on chromosomes 2, 6, 14 and 20 in Norwegian red cattle. Anim Genet 42:457–465. https://doi.org/10.1111/j.1365-2052.2010.02165.x
Souza FN, Cunha AF, Rosa DLSO, Brito MAV, Guimarães AS, Mendonça LC, Souza GN, Lage AP, Blagitz MG, Libera AMMPD, Heinemann MB, Cerqueira MMOP (2016) Somatic cell count and mastitis pathogen detection in composite and single or duplicate quarter milk samples. Pesqui Vet Bras 36:811–818. https://doi.org/10.1590/S0100-736X2016000900004
Suriyasathaporn W, Schukken YH, Nielen M, Brand A (2000) Low somatic cell count: a risk factor for subsequent clinical mastitis in a dairy herd. J Dairy Sci 83:1248–1255. https://doi.org/10.3168/jds.S0022-0302(00)74991-5
Tiezzil F, Parker-Gaddis KL, Cole JB et al (2015) A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS One 10:e0114919. https://doi.org/10.1371/journal.pone.0114919
Waage S, Sviland S, Ødegaard SA (1998) Identification of risk factors for clinical mastitis in dairy heifers. J Dairy Sci 81:1275–1284. https://doi.org/10.3168/jds.S0022-0302(98)75689-9
Wu X, Lund MS, Sahana G, Guldbrandtsen B, Sun D, Zhang Q, Su G (2015) Association analysis for udder health based on SNP-panel and sequence data in Danish Holsteins. Genet Sel Evol 47:50. https://doi.org/10.1186/s12711-015-0129-1
Ziegler A, König IR, Thompson JR (2008) Biostatistical aspects of genome-wide association studies. Biom J 50:8–28. https://doi.org/10.1002/bimj.200710398
Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, van Tassell CP, Sonstegard TS, Marçais G, Roberts M, Subramanian P, Yorke JA, Salzberg SL (2009) A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol 10:R42. https://doi.org/10.1186/gb-2009-10-4-r42
Acknowledgments
The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged.
Funding
Funding for sample collection and data analysis was provided by the Utah Agriculture Experiment Station (Utah State University Extension Grants Program to Zhongde Wang) and the Utah Department of Agriculture and Food (Cap Ferry Agricultural Grant Fund to Zhongde Wang). The funding body had no role in the design of the study and collection, analysis, and interpretation of data or in writing the manuscript. The computational resources used were partially funded by the NIH Shared Instrumentation Grant 1S10OD021644-01A1.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The use of animals in this study was approved by the Utah State University Institutional Animal Care and Use Committee (protocol IACUC-2282), and permission was obtained from the cattle owner. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practices at which the studies were conducted.
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Online Resource 1
SNPs nominal for genome-wide significance for bovine mastitis resistance. (XLSX 43 kb)
Rights and permissions
About this article
Cite this article
Kurz, J.P., Yang, Z., Weiss, R.B. et al. A genome-wide association study for mastitis resistance in phenotypically well-characterized Holstein dairy cattle using a selective genotyping approach. Immunogenetics 71, 35–47 (2019). https://doi.org/10.1007/s00251-018-1088-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-018-1088-9