Abstract
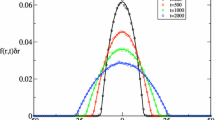
Fluorescence correlation spectroscopy was used to measure the diffusion behavior of a mixture of DMPC or DMPC/DMPG liposomes with human serum albumin (HSA) and mesoporphyrin (MP), which was used as the fluorescent label for liposomes and HSA as well. For decomposing the fluorescence intensity autocorrelation function (ACF) into components corresponding to a liposome population, HSA and MP, we used a maximum entropy procedure that computes a distribution of diffusion times consistent with the ACF data. We found that a simple parametric non-linear fit with a discrete set of decay components did not converge to a stable parameter set. The distribution calculated with the maximum entropy method was stable and the average size of the particles calculated from the effective diffusion time was in good agreement with the data determined using the discrete-component fit.





Similar content being viewed by others
References
Bárdos-Nagy I, Galantai R, Kaposi A, Fidy J (1998) Difference in the transport of metal and free-base porphyrins. Steady-state and time-resolved fluorescence studies. Int J Pharm 175:255–267
Bardos-Nagy I, Galantai R, Laberge M, Fidy J (2003) The effect of trehalose on the nonbond associative interactions between small unilamellar vesicles and human serum albumin and on the aging process. Langmuir 19:146–153
Bryan R (1990) Maximum entropy analysis of oversampled data problems. Eur Biophys J 18:165
Chen Y, Müller JD, Tetin SY, Tyner JD, Gratton E (2000) Probing ligand protein binding equilibria with fluorescence fluctuation spectroscopy. Biophys J 79:1074–1084
Elson EL, Magde D (1974) Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers 13:1–27
Ferrer ML, Duchowicz R, Carrasco B, de la Torre, JG, Acuna AU (2001) The conformation of serum albumin in solution: a combined phosphorescence depolarization-hydrodynamic modeling study. Biophys J 80:2422–2430
Galantai R, Bardos-Nagy I (2000) The interaction of human serum albumin and model membranes. Int J Pharm 195:207–18
Galantai R, Bardos-Nagy I, Modos K, Kardos J, Zavodszky P, Fidy J (2000) Serum albumin-lipid membrane interaction influencing the uptake of porphyrins. Arch Biochem Biophys 373:261–270
Gull SF, Daniell GJ (1978) Image reconstruction from incomplete and noisy data. Nature 272:686–690
Langowski J, Bryan R (1991) Maximum entropy analysis of photon correlation spectroscopy data using a Bayesian estimate for the regularization parameter. Macromolecules 24:6346–6348
Langowski J, Tewes M (2000) Determination of DNA-ligand interactions by fluorescence correlation spectroscopy. In: Travers A, Buckle M (eds) Protein-DNA interactions: a practical approach. Oxford University Press, Oxford, pp 95–111
Magde D, Elson EL, Webb WW (1972) Thermodynamic fluctuations in a reacting system—measurement by fluorescence correlations spectroscopy. Phys Rev Lett 29:705–708
Magde D, Elson EL, Webb WW (1974) Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers 13:29–61
Oncley JL, Scatchard G, Brown A (1947) Physical-chemical characteristics of certain of the proteins of normal human plasma. J Phys Colloid Chem 51:184
Press WH, Flannery BP, Teukolsky SA, Vetterling VT (1986) Numerical recipes in C: the art of scientific computing. Cambridge University Press, Cambridge
Qian H, Elson EL (1991) Analysis of confocal laser-microscope optics for 3-D fluorescence correlation spectroscopy. Appl Opt 30:1185–1195
Reddi E, Ricchelli F, Jori G (1981) Interaction of human serum albumin with hematoporphyrin and its Zn(2)+- and Fe(3)+-derivatives. Int J Pept Protein Res 18:402–408
Rigler R, Mets Ü, Widengren J, Kask P (1993) Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. Eur Biophys J 22:169–175
Rouser G, Simon G, Kritchevsky G (1969) Species variations in phospholipid class distribution of organs. I. Kidney, liver and spleen. Lipids 4:599–606
Skilling J, Bryan RK (1984) Maximum entropy image reconstruction: general algorithm. Mon Not R Astron Soc 211:111–124
Widengren J, Mets Ü, Rigler R (1995) Fluorescence correlation spectroscopy of triplet states in solution: a theoretical and experimental study. J Phys Chem 99:13368–13379
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Módos, K., Galántai, R., Bárdos-Nagy, I. et al. Maximum-entropy decomposition of fluorescence correlation spectroscopy data: application to liposome–human serum albumin association. Eur Biophys J 33, 59–67 (2004). https://doi.org/10.1007/s00249-003-0343-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-003-0343-6