Abstract
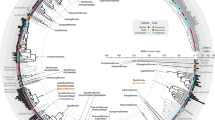
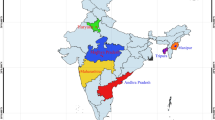
Intestinal microbiota plays an important role in promoting digestion, metabolism, and immunity. Intestinal microbiota and fatty acids are important indicators to evaluate the health and nutritional composition of Procambarus clarkii. They have been shown to be strongly influence by environmental and genetic factors. However, it is not clear whether environmental factors have a greater impact on the intestinal microbiota and fatty acid composition of crayfish. The link between the intestinal microbial communities and fatty acid (FA) compositions of red swamp crayfish from different geographical has not yet been studied. Thus, the current paper focuses on the influence of different environments on the fatty acids in muscles of crayfish and the possible existence between gut microbiota and fatty acids. Therefore, in this study, we compared the fatty acid compositions and intestinal microbiota of five crayfish populations from different geographical locations. The results were further analyzed to determine whether there is a relationship between geographical location, fatty acid compositions and intestinal microbiota. The gut microbial communities of the crayfish populations were characterized using 16S rRNA high-throughput gene sequencing. The results showed that there were significant differences in FA compositions of crayfish populations from different geographical locations. A similar trend was observed in the gut microbiome, which also varied significantly according to geographic location. Interestingly, the analysis revealed that there was a relationship between fatty acid compositions and intestinal microbes, revealed by alpha diversity analysis and cluster analysis. However, further studies of the interactions between the P. clarkii gut microbiota and biochemical composition are needed, which will ultimately reveal the complexity of microbial ecosystems with potential applications in aquaculture and species conservation.






Similar content being viewed by others
Data Availability
The datasets presented in this study are available in online repositories. The names of the repository/repositories and accession number(s) can be found below. https://dataview.ncbi.nlm.nih.gov/object/PRJNA1018237.
References
Li J, Chen JH, Li L, Li J (2021) Evaluation of the nutritional quality of edible tissues (muscle and hepatopancreas) of cultivated Procambarus clarkii using biofloc technology. Aquac Rep 19:100586. https://doi.org/10.1016/j.aqrep.2021.100586
Du Zhi-Qiang LX-Y, Xiu-Li S, Yan-Hui J, Xin-Cang L (2017) Identification and characterization of lymph organ microRNAs in red swamp crayfish, Procambarus clarkii infected with white spot syndrome virus. Fish Shellfish Immunol:78–84. https://doi.org/10.1016/j.fsi.2017.08.007
Tian J, Qiao-Qing XU, Tian L, Wei HU, Yang CG, Gao WH (2017) The muscle composition analysis and flesh quality of procambarus clarkia in the dongting lake. Acta Hydrobiol Sin. https://doi.org/10.7541/2017.108
Fan L, Li F, Chen X, Qiu L, Chen J (2021) Metagenomics analysis reveals the distribution and communication of antibiotic resistance genes within two different red swamp crayfish Procambarus clarkii cultivation ecosystems. Environmental Pollution 285:117144. https://doi.org/10.1016/j.envpol.2021.117144
Li YS, Liu QG, Chen LS (2008) Ecological culture of Procambarus clarkii Artificial breeding and rearing of 2 gram Procambarus clarkii. Aquat Sci Technol Inform 251:84–87
Ross BD, Verster AJ, Radey MC, Schmidtke DT, Mougous JD (2019) Human gut bacteria contain acquired interbacterial defence systems. Nature 575. https://doi.org/10.1038/s41586-019-1708-z
Su S, Munganga BP, Du F, Yu J, Li J, Yu F, Wang M, He X, Li X, Bouzoualegh R, Xu P, Tang Y (2020) Relationship Between the Fatty Acid Profiles and Gut Bacterial Communities of the Chinese Mitten Crab (Eriocheir sinensis) From Ecologically Different Habitats. Front Microbiol 11:565267. https://doi.org/10.3389/fmicb.2020.565267
Eisenstein M (2020) The hunt for a healthy microbiome. Nature 577:S6–S8. https://doi.org/10.1038/d41586-020-00193-3
Cho I, Blaser MJ (2012) The human microbiome: at the interface of health and disease. Nat Rev Genet:260–270. https://doi.org/10.1038/nrg3182
Hooper LV, Dan RL, Macpherson AJ (2012) Interactions Between the Microbiota and the Immune System. Science 336:1268–1273. https://doi.org/10.1126/science.1223490
Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, Sinha R, Peterson DA, Walter J, Benson AK, Pomp D (2014) Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol 15:552. https://doi.org/10.1186/s13059-014-0552-6
Bascuñán P, Niño-Garcia JP, Galeano-Castañeda Y, Serre D, Correa MM (2018) Factors shaping the gut bacterial community assembly in two main Colombian malaria vectors. Microbiome 6:148. https://doi.org/10.1186/s40168-018-0528-y
Contreras M, Magris M, Hidalgo G, Robert N, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG (2012) Human gut microbiome viewed across age and geography. Nature 486:222–227. https://doi.org/10.1038/nature11053
Zhang M, Sun Y, Chen L, Cai C, Qiao F, Du Z, Li E (2016) Symbiotic Bacteria in Gills and Guts of Chinese Mitten Crab (Eriocheir sinensis) Differ from the Free-Living Bacteria in Water. Plos One 11:e0148135. https://doi.org/10.1371/journal.pone.0148135
Feng, Y.X., Feng, N.X., and Chen, X. (2020) Interaction between Environmental Pollutants and Gut Microbiota: A Review
Rothschild D, Weissbrod O, Barkan E et al (2018) Environment dominates over host genetics in shaping human gut microbiota. Nature 555:210–215. https://doi.org/10.1038/nature25973
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Applenvironmicrobiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
White JR, Nagarajan N, Pop M (2009) Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLOS Comput Biol 5. https://doi.org/10.1371/journal.pcbi.1000352
Li B, Zhang X, Guo F, Wu W, Zhang T (2013) Characterization of tetracycline resistant bacterial community in saline activated sludge using batch stress incubation with high-throughput sequencing analysis. Water Res 47:4207–4216. https://doi.org/10.1016/j.watres.2013.04.021
Alvanou MV, Feidantsis K, Staikou A, Apostolidis AP, Michaelidis B, Giantsis IA (2023) Probiotics, Prebiotics, and Synbiotics Utilization in Crayfish Aquaculture and Factors Affecting Gut Microbiota. Microorganisms 11. https://doi.org/10.3390/microorganisms11051232
Sang L (2009) The impact of water environment on the fatty acids of Lhasa Naked Carpin. J Tibet Univ
Rutkowska J, Bialek M, Adamska A, Zbikowska A (2015) Differentiation of geographical origin of cream products in Poland according to their fatty acid profile. Food Chem 178:26–31. https://doi.org/10.1016/j.foodchem.2015.01.036
Han N, Wang Y, Li MA, Shi XX, Liu ZZ, Zhang SG (2018) Determination and Study of Fatty Acid and Amino Acid in Lamb on Three Different Regions of Gansu Province. Food Ferment Sci Technol
Sillero-Ríos J, Sureda A, Capó X, Oliver-Codorniú M, Arechavala-Lopez P (2018) Biomarkers of physiological responses of Octopus vulgaris to different coastal environments in the western Mediterranean Sea. Mar Pollut Bull 128:240. https://doi.org/10.1016/j.marpolbul.2018.01.032
Cai LP, Hai Sheng XU, Lin HE, Shu MA (2011) Analysis of the intestinal bacterial communities in wild Scylla serrata from different districts by PCR-DGGE. Acta Agriculturae Zhejiangensis 23:278–282. https://doi.org/10.1016/S1671-2927(11)60313-1
Shui Y, Guan Z-B, Liu G-F, Fan L-M (2020) Gut microbiota of red swamp crayfish Procambarus clarkii in integrated crayfish-rice cultivation model. AMB Express 10:5. https://doi.org/10.1186/s13568-019-0944-9
Ludek Sehnal, Elizabeth Brammer-Robbins, Alexis M. Wormington, Ludek Blaha, Ondrej Adamovsky. (2021) Microbiome Composition and Function in Aquatic Vertebrates: Small Organisms Making Big Impacts on Aquatic Animal Health. Front Microbiol 12: 567408. https://doi.org/10.3389/fmicb.2021.567408
Pedà C, Caccamo L, Fossi MC, Gai F, Andaloro F, Genovese L, Perdichizzi A, Romeo T, Maricchiolo G (2016) Intestinal alterations in European sea bass Dicentrarchus labrax (Linnaeus, 1758) exposed to microplastics: Preliminary results. Environ Pollut 212:251–256. https://doi.org/10.1016/j.scitotenv.2021.148224
I Brila, A Lavrinienko, E Tukalenko, F Ecke, I Rodushkin, ER Kallio, T Mappes, PC Watts. (2021) Low-level environmental metal pollution is associated with altered gut microbiota of a wild rodent, the bank vole (Myodes glareolus). Sci Total Environ 790: 148224. https://doi.org/10.1016/j.scitotenv.2021.148224
Yuhui L, Ting C, Youbang L, Yin T, Zhonghao H (2021) Gut microbiota are associated with sex and age of host: Evidence from semi-provisioned rhesus macaques in southwest Guangxi, China. Ecology and Evolution, p 11
Zhang Z, Chen X, Loh YJ, Yang X, Zhang C The effect of calorie intake, fasting, and dietary composition on metabolic health and gut microbiota in mice. BMC Biol 19:51. https://doi.org/10.1186/s12915-021-00987-5
Zhang Z, Liu J, Jin X, Liu C, Fan C, Guo L, Liang Y, Zheng J, Peng N (2020) Developmental, Dietary, and Geographical Impacts on Gut Microbiota of Red Swamp Crayfish (Procambarus clarkii). Microorganisms 8. https://doi.org/10.3390/microorganisms8091376
Fan J, Chen L, Mai G, Zhang H, Yang J, Deng D, Ma Y (2019) Dynamics of the gut microbiota in developmental stages of Litopenaeus vannamei reveal its association with body weight. Sci Rep 9. https://doi.org/10.1038/s41598-018-37042-3
Qin J, Li R, Raes J et al (2010) A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. https://doi.org/10.1038/nature08821
Kostanjsek R, Strus J, Avgustin G (2007) "Candidatus Bacilloplasma," a Novel Lineage of Mollicutes Associated with the Hindgut Wall of the Terrestrial Isopod Porcellio scaber (Crustacea: Isopoda). Appl Environ Microbiol 73:5566–5573. https://doi.org/10.1128/AEM.02468-06
Acknowledgements
We thank the students and staff of the Aquatic Genetic Laboratory, FFRC for their assistance in this study.
Funding
This work was supported by grants from the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS [2021]123), Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2022XT01), Central Public-interest Scientific Institution Basal Research Fund, Freshwater Fisheries Research Center, CAFS (NO. 2023JBFM21).
Author information
Authors and Affiliations
Contributions
Shengyan Su conceived the study and contributed to the design of the experiments. Ming Chen, Wenrong Feng, Can Tian, Jingjing Jiang, Yang Gao, Zeyu Lu, Jian zhu, Chengfeng Zhang performed all the experiments. All authors contributed to the drafting of the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, M., Su, S., Zhang, C. et al. The Role of Biogeography in Shaping Intestinal Flora and Influence on Fatty Acid Composition in Red Swamp Crayfish (Procambarus clarkii). Microb Ecol 86, 3111–3127 (2023). https://doi.org/10.1007/s00248-023-02298-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-023-02298-4