Abstract
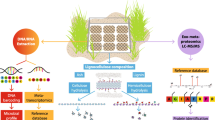
Terrestrial microbial consortia were reported to play fundamental roles in the global carbon cycle and renewable energy production through the breakdown of complex organic carbon. However, we have a poor understanding of how biotic/abiotic factors combine to influence consortia assembly and lignocellulose degradation in aquatic ecosystems. In this study, we used 96 in situ lignocellulose enriched, coastal intertidal zone-derived bacterial consortia as the initial inoculating consortia and developed 384 cultured consortia under different lignocellulose substrates (aspen, pine, rice straw, and purified Norway spruce lignin) with gradients of salinity and temperature. As coastal consortia, salinity was the strongest driver for assembly, followed by Norway spruce lignin, temperature, and aspen. Moreover, a conceptual model was proposed to demonstrate different succession dynamics between consortia under herbaceous and woody lignocelluloses. The succession of consortium under Norway spruce lignin is greatly related with abiotic factors, while its substrate degradation is mostly correlated with biotic factors. A discrepant pattern was observed in the consortium under rice straw. Finally, we developed four groups of versatile, yet specific consortia. Our study not only reveals that coastal intertidal wetlands are important natural resources to enrich lignocellulolytic degrading consortia but also provides insights into the succession and ecological function of coastal consortium.





Similar content being viewed by others
Data Availability
All sequencing data have been deposited in the NCBI SRA database under the accession number PRJNA865006.
References
Becker J, Wittmann C (2019) A field of dreams: lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol Adv 37:107360. https://doi.org/10.1016/j.biotechadv.2019.02.016
Lin L, Wang X, Cao L, Xu M (2019) Lignin catabolic pathways reveal unique characteristics of dye-decolorizing peroxidases in Pseudomonas putida. Environ Microbiol 21:1847–1863. https://doi.org/10.1111/1462-2920.14593
Lin L (2022) Bottom-up synthetic ecology study of microbial consortia to enhance lignocellulose bioconversion. Biotechnol Biofuels Bioprod 15:14. https://doi.org/10.1186/s13068-022-02113-1
Peng X, Wilken SE, Lankiewicz TS, Gilmore SP, Brown JL, Henske JK, Swift CL, Salamov A, Barry K, Grigoriev IV, Theodorou MK, Valentine DL, O’Malley MA (2021) Genomic and functional analyses of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat Microbiol 6:499–511. https://doi.org/10.1038/s41564-020-00861-0
Díaz-García L, Huang S, Spröer C, Sierra-Ramírez R, Bunk B, Overmann J, Jiménez DJ (2021) Dilution-to-stimulation/extinction method: a combination enrichment strategy to develop a minimal and versatile lignocellulolytic bacterial consortium. Appl Environ Microbiol 87:e02427-e2420. https://doi.org/10.1128/aem.02427-20
Cortes-Tolalpa L, Jiménez DJ, de Lima Brossi MJ, Salles JF, van Elsas JD (2016) Different inocula produce distinctive microbial consortia with similar lignocellulose degradation capacity. Appl Microbiol Biotechnol 100:7713–7725. https://doi.org/10.1007/s00253-016-7516-6
Zegeye EK, Brislawn CJ, Farris Y, Fansler SJ, Hofmockel KS, Jansson JK, Wright AT, Graham EB, Naylor D, McClure RS, Bernstein HC (2019) Selection, succession, and stabilization of soil microbial consortia. mSystems 4. https://doi.org/10.1128/mSystems.00055-19
Kolinko S, Wu YW, Tachea F, Denzel E, Hiras J, Gabriel R, Bäcker N, Chan LJG, Eichorst SA, Frey D, Chen Q, Azadi P, Adams PD, Pray TR, Tanjore D, Petzold CJ, Gladden JM, Simmons BA, Singer SW (2018) A bacterial pioneer produces cellulase complexes that persist through community succession. Nat Microbiol 3:99–107. https://doi.org/10.1038/s41564-017-0052-z
Wang W, Zhang Q, Sun X, Chen D, Insam H, Koide RT, Zhang S (2020) Effects of mixed-species litter on bacterial and fungal lignocellulose degradation functions during litter decomposition. Soil Biol Biochem 141:107690. https://doi.org/10.1016/j.soilbio.2019.107690
Fang X, Li Q, Lin Y, Lin X, Dai Y, Guo Z, Pan D (2018) Screening of a microbial consortium for selective degradation of lignin from tree trimmings. Bioresour Technol 254:247–255. https://doi.org/10.1016/j.biortech.2018.01.058
Zhang G, Bai J, Tebbe CC, Zhao Q, Jia J, Wang W, Wang X, Yu L (2021) Salinity controls soil microbial community structure and function in coastal estuarine wetlands. Environ Microbiol 23:1020–1037. https://doi.org/10.1111/1462-2920.15281
Osland MJ, Enwright NM, Day RH, Gabler CA, Stagg CL, Grace JB (2016) Beyond just sea-level rise: considering macroclimatic drivers within coastal wetland vulnerability assessments to climate change. Glob Chang Biol 22:1–11. https://doi.org/10.1111/gcb.13084
Zhang Z, Han P, Zheng Y, Jiao S, Dong H, Liang X, Gao D, Niu Y, Yin G, Liu M, Hou L (2022) Spatiotemporal dynamics of bacterial taxonomic and functional profiles in estuarine intertidal soils of China coastal zone. Microb Ecol. https://doi.org/10.1007/s00248-022-01996-9
Puentes-Téllez PE, Salles JF (2020) Dynamics of abundant and rare bacteria during degradation of lignocellulose from sugarcane biomass. Microb Ecol 79:312–325. https://doi.org/10.1007/s00248-019-01403-w
Pérez-Hernández V, Hernández-Guzmán M, Luna-Guido M, Navarro-Noya YE, Romero-Tepal EM, Dendooven L (2021) Bacterial communities in alkaline saline soils amended with young maize plants or its (hemi)cellulose fraction. Microorganisms 9:1297. https://doi.org/10.3390/microorganisms9061297
Tláskal V, Brabcová V, Větrovský T, Jomura M, López-Mondéjar R, Oliveira Monteiro LM, Saraiva JP, Human ZR, Cajthaml T, Nunes da Rocha U, Baldrian P (2021) Complementary roles of wood-inhabiting fungi and bacteria facilitate deadwood decomposition.mSystems 6. https://doi.org/10.1128/mSystems.01078-20
Haq IU, Hillmann BM, Moran MK, Willard S, Knights D, Fixen KR, Schilling JS (2022) Bacterial communities associated with wood rot fungi that use distinct decomposition mechanisms. ISME Commun. https://doi.org/10.1038/s43705-022-00108-5
Zhang J, Zhang W, Cai Z, Zhang J, Guan D, Ji D, Gao W (2022) Effect of ammonia fiber expansion combined with NaOH pretreatment on the resource efficiency of herbaceous and woody lignocellulosic biomass. ACS Omega 7:18761–18769. https://doi.org/10.1021/acsomega.2c01302
Cortes-Tolalpa L, Norder J, van Elsas JD, Falcao Salles J (2018) Halotolerant microbial consortia able to degrade highly recalcitrant plant biomass substrate. Appl Microbiol Biotechnol 102:2913–2927. https://doi.org/10.1007/s00253-017-8714-6
Wang Y, Elzenga T, van Elsas JD (2021) Effect of culture conditions on the performance of lignocellulose-degrading synthetic microbial consortia. Appl Microbiol Biotechnol 105:7981–7995. https://doi.org/10.1007/s00253-021-11591-6
Leadbeater DR, Oates NC, Bennett JP, Li Y, Dowle AA, Taylor JD, Alponti JS, Setchfield AT, Alessi AM, Helgason T, McQueen-Mason SJ, Bruce NC (2021) Mechanistic strategies of microbial communities regulating lignocellulose deconstruction in a UK salt marsh. Microbiome 9:48. https://doi.org/10.1186/s40168-020-00964-0
Cragg SM, Beckham GT, Bruce NC, Bugg TD, Distel DL, Dupree P, Etxabe AG, Goodell BS, Jellison J, McGeehan JE, McQueen-Mason SJ, Schnorr K, Walton PH, Watts JE, Zimmer M (2015) Lignocellulose degradation mechanisms across the Tree of Life. Curr Opin Chem Biol 29:108–119. https://doi.org/10.1016/j.cbpa.2015.10.018
Meslé MM, Mueller RC, Peach J, Eilers B, Tripet BP, Bothner B, Copié V, Peyton BM (2022) Isolation and characterization of lignocellulose-degrading Geobacillus thermoleovorans from Yellowstone National Park. Appl Environ Microbiol 88:e0095821. https://doi.org/10.1128/aem.00958-21
Tomazetto G, Pimentel AC, Wibberg D, Dixon N, Squina FM (2020) Multi-omic directed discovery of cellulosomes, polysaccharide utilization loci, and lignocellulases from an enriched rumen anaerobic consortium. Appl Environ Microbiol 86:e00199-e120. https://doi.org/10.1128/aem.00199-20
Wang X, Lin L, Zhou J (2021) Links among extracellular enzymes, lignin degradation and cell growth establish the models to identify marine lignin-utilizing bacteria. Environ Microbiol 23:160–173. https://doi.org/10.1111/1462-2920.15289
Wang X, Lin L, Dong J, Ling J, Wang W, Wang H, Zhang Z, Yu X (2018) Simultaneous improvements of Pseudomonas cell growth and polyhydroxyalkanoate production from a lignin derivative for lignin-consolidated bioprocessing. Appl Environ Microbiol 84:e01469-e1418. https://doi.org/10.1128/aem.01469-18
Carroll J, Van Oostende N, Ward BB (2022) Evaluation of genomic sequence-based growth rate methods for Synchronized Synechococcus cultures. Appl Environ Microbiol 88:e0174321. https://doi.org/10.1128/aem.01743-21
Cao L, Lin L, Sui H, Wang H, Zhang Z, Jiao N, Zhou J (2021) Efficient extracellular laccase secretion via bio-designed secretory apparatuses to enhance bacterial utilization of recalcitrant lignin. Green Chem 23:2079–2094. https://doi.org/10.1039/D0GC04084C
Singh A, Kumar R, Maurya A, Chowdhary P, Raj A (2022) Isolation of functional ligninolytic Bacillus aryabhattai from paper mill sludge and its lignin degradation potential. Biotechnol Rep (Amst) 35:e00755. https://doi.org/10.1016/j.btre.2022.e00755
Xu C, Su X, Wang J, Zhang F, Shen G, Yuan Y, Yan L, Tang H, Song F, Wang W (2021) Characteristics and functional bacteria in a microbial consortium for rice straw lignin-degrading. Bioresour Technol 331:125066. https://doi.org/10.1016/j.biortech.2021.125066
Brown ME, Walker MC, Nakashige TG, Iavarone AT, Chang MC (2011) Discovery and characterization of heme enzymes from unsequenced bacteria: application to microbial lignin degradation. J Am Chem Soc 133:18006–18009. https://doi.org/10.1021/ja203972q
Brossi MJdL, Jiménez DJ, Cortes-Tolalpa L, Elsas JD (2016) Soil-derived microbial consortia enriched with different plant biomass reveal distinct players acting in lignocellulose degradation. Microb Ecol 71:616–627. https://doi.org/10.1007/s00248-015-0683-7
Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D (2012) Determination of structural carbohydrates and lignin in biomass. NREL Lab Anal Proced (LAP)
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/aem.00062-07
Du S, Dini-Andreote F, Zhang N, Liang C, Yao Z, Zhang H, Zhang D (2020) Divergent co-occurrence patterns and assembly processes structure the abundant and rare bacterial communities in a salt marsh ecosystem. Appl Environ Microbiol 86:e00322-e320. https://doi.org/10.1128/aem.00322-20
Zhou J, Liu W, Deng Y, Jiang YH, Xue K, He Z, Van Nostrand JD, Wu L, Yang Y, Wang A (2013) Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio 4:e00584-00512. https://doi.org/10.1128/mBio.00584-12
Wang S, Xiong W, Wang Y, Nie Y, Wu Q, Xu Y, Geisen S (2020) Temperature-induced annual variation in microbial community changes and resulting metabolome shifts in a controlled fermentation system. mSystems 5:e00555-00520. https://doi.org/10.1128/mSystems.00555-20
Oksanen J, Simpson G, Blanchet FG, Kindt R, Legendre P, Minchin P, hara R, Solymos P, Stevens H, Szöcs E, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, De Cáceres M, Durand S, Weedon J (2022) Vegan community ecology package version 2.6–2 April 2022
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. https://doi.org/10.1186/gb-2011-12-6-r60
Kendall MG (1945) The treatment of ties in ranking problems. Biometrika 33:239–251. https://doi.org/10.1093/biomet/33.3.239
Song W, Liu J, Qin W, Huang J, Yu X, Xu M, Stahl D, Jiao N, Zhou J, Tu Q (2022) Functional traits resolve mechanisms governing the assembly and distribution of nitrogen-cycling microbial communities in the global ocean. mBio 13:e0383221. https://doi.org/10.1128/mbio.03832-21
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2:18–22
Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38:685–688. https://doi.org/10.1038/s41587-020-0548-6
Farda B, Vaccarelli I, Ercole C, Djebaili R, Del Gallo M, Pellegrini M (2022) Exploring structure, microbiota, and metagenome functions of epigean and hypogean black deposits by microscopic, molecular and bioinformatics approaches. Sci Rep 12:19405. https://doi.org/10.1038/s41598-022-24159-9
Jiménez DJ, de Lima Brossi MJ, Schückel J, Kračun SK, Willats WG, van Elsas JD (2016) Characterization of three plant biomass-degrading microbial consortia by metagenomics- and metasecretomics-based approaches. Appl Microbiol Biotechnol 100:10463–10477. https://doi.org/10.1007/s00253-016-7713-3
Ning D, Deng Y, Tiedje JM, Zhou J (2019) A general framework for quantitatively assessing ecological stochasticity. Proc Natl Acad Sci U S A 116:16892–16898. https://doi.org/10.1073/pnas.1904623116
Mathé C, Fawal N, Roux C, Dunand C (2019) In silico definition of new ligninolytic peroxidase sub-classes in fungi and putative relation to fungal life style. Sci Rep 9:20373. https://doi.org/10.1038/s41598-019-56774-4
Ruiz-Dueñas FJ, Martínez AT (2009) Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb Biotechnol 2:164–177. https://doi.org/10.1111/j.1751-7915.2008.00078.x
Lewin GR, Davis NM, McDonald BR, Book AJ, Chevrette MG, Suh S, Boll A, Currie CR (2022) Long-term cellulose enrichment selects for highly cellulolytic consortia and competition for public goods. mSystems 7: e0151921. https://doi.org/10.1128/msystems.01519-21
Hui W, Jiajia L, Yucai L, Peng G, Xiaofen W, Kazuhiro M, Zongjun C (2013) Bioconversion of un-pretreated lignocellulosic materials by a microbial consortium XDC-2. Bioresour Technol 136:481–487. https://doi.org/10.1016/j.biortech.2013.03.015
Cheng Y, Huang M, Shen X, Jiang C (2022) Enhanced cornstalk decomposition by a psychrotrophic bacterial consortium comprising cellulose, hemicellulose, and lignin degraders with biochar as a carrier for carbonneutrality. Bioresour Technol 344:126259. https://doi.org/10.1016/j.biortech.2021.126259
Lu J, Yang Z, Xu W, Shi X, Guo R (2019) Enrichment of thermophilic and mesophilicmicrobial consortia for efficient degradation of corn stalk. J Environ Sci (China) 78:118–126. https://doi.org/10.1016/j.jes.2018.07.010
Zhang D, Wang Y, Zhang C, Zheng D, Guo P, Cui Z (2018) Characterization of a thermophilic lignocellulose-degrading microbial consortium with high extracellular xylanase activity. J Microbiol Biotechnol 28:305–313. https://doi.org/10.4014/jmb.1709.09036
Tahir AA, Mohd Barnoh NF, Yusof N, Mohd Said NN, Utsumi M, Yen AM, Hashim H, Mohd Noor MJM, Akhir FNM, Mohamad SE, Sugiura N, Othman N, Zakaria Z, Hara H (2019) Microbial diversity in decaying oil palm empty fruit bunches (opefb) and isolation of lignin-degrading bacteria from a tropical environment. Microbes Environ 34:161–168. https://doi.org/10.1264/jsme2.ME18117
Ohta Y, Nishi S, Haga T, Tsubouchi T, Hasegawa R, Konishi M, Nagano Y, Tsuruwaka Y, Shimane Y, Mori K, Usui K, Suda E, Tsutsui K, Nishimoto A, Fujiwara Y, Maruyama T, Hatada Y (2012) Screening and phylogenetic analysis of deep-sea bacteria capable of metabolizing lignin-derived aromatic compounds. OJMS 2:177–187. https://doi.org/10.4236/ojms.2012.24021
Hu Y, Guo Y, Lai Q, Dong L, Huang Z (2020) Draconibacterium mangrovi sp. nov., isolated from mangrove sediment. Int J Syst Evol Microbiol 70:4816–4821. https://doi.org/10.1099/ijsem.0.004354
Zhou J, Ning D (2017) Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81. https://doi.org/10.1128/mmbr.00002-17
Tripathi S, Sharma P, Chandra R (2021) Degradation of organometallic pollutants of distillery wastewater by autochthonous bacterial community in biostimulation and bioaugmentation process. Bioresour Technol 338:125518. https://doi.org/10.1016/j.biortech.2021.125518
Puentes-Téllez PE, Falcao Salles J (2018) Construction of effective minimal active microbial consortia for lignocellulose degradation. Microb Ecol 76:419–429. https://doi.org/10.1007/s00248-017-1141-5
Meng L, Xu C, Wu F, Huhe (2022) Microbial co-occurrence networks driven by low-abundance microbial taxa during composting dominate lignocellulose degradation. Sci Total Environ 845: 157197. https://doi.org/10.1016/j.scitotenv.2022.157197
Morrissey EM, Gillespie JL, Morina JC, Franklin RB (2014) Salinity affects microbial activity and soil organic matter content in tidal wetlands. Glob Chang Biol 20:1351–1362. https://doi.org/10.1111/gcb.12431
Xia S, Song Z, Li Q, Guo L, Yu C, Singh BP, Fu X, Chen C, Wang Y, Wang H (2021) Distribution, sources, and decomposition of soil organic matter along a salinity gradient in estuarine wetlands characterized by C: N ratio, δ(13) C-δ(15) N, and lignin biomarker. Glob Chang Biol 27:417–434. https://doi.org/10.1111/gcb.15403
Oates NC, Abood A, Schirmacher AM, Alessi AM, Bird SM, Bennett JP, Leadbeater DR, Li Y, DowleAA, Liu S, Tymokhin VI, Ralph J, McQueen-Mason SJ, Bruce NC (2021) A multi-omics approach to lignocellulolytic enzyme discovery reveals a new ligninase activity from Parascedosporium putredinis NO1. Proc Natl Acad Sci U S A 118. https://doi.org/10.1073/pnas.2008888118
Xun W, Li W, Xiong W, Ren Y, Liu Y, Miao Y, Xu Z, Zhang N, Shen Q, Zhang R (2019) Diversity-triggered deterministic bacterial assembly constrains community functions. Nat Commun 10:3833. https://doi.org/10.1038/s41467-019-11787-5
Funding
This work was funded by the National Natural Science Foundation of China (91951116) and National Key Research and Development Project (2019YFA0606704).
Author information
Authors and Affiliations
Contributions
LL conceived the study. MWW performed the experiments and analyzed the data. PQN performed in situ enrichment experiment and assisted the data analysis. LL interpreted the data and wrote the manuscript with input from MWW. All authors approved the final manuscript and declare no conflict of interest.
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, W., Lin, L. & Peng, Q. Origin, Selection, and Succession of Coastal Intertidal Zone-Derived Bacterial Communities Associated with the Degradation of Various Lignocellulose Substrates. Microb Ecol 86, 1589–1603 (2023). https://doi.org/10.1007/s00248-023-02170-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-023-02170-5