Abstract
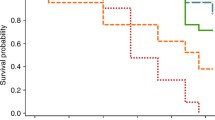
Host genotype and environment are considered crucial factors in shaping Daphnia gut microbiome composition. Among the environmental factors, diet is an important factor that regulates Daphnia microbiome. Most of the studies only focused on the use of axenic diet and non-sterile medium to investigate their effects on Daphnia microbiome. However, in natural environment, Daphnia diets such as phytoplankton are associated with microbes and could affect Daphnia microbiome composition and fitness, but remain relatively poorly understood compared to that of axenic diet. To test this, we cultured two Daphnia magna genotypes (genotype-1 and genotype-2) in sterile medium and fed with axenic diet. To check the effects of algal diet-associated microbes versus free water-related microbes, Daphnia were respectively inoculated with three different inoculums: medium microbial inoculum, diet-associated microbial inoculum, and medium and diet-mixed microbial inoculum. Daphnia were cultured for 3 weeks and their gut microbiome and life history traits were recorded. Results showed that Daphnia inoculated with medium microbial inoculum were dominated by Comamonadaceae in both genotypes. In Daphnia inoculated with mixed inoculum, genotype-1 microbiome was highly changed, whereas genotype-2 microbiome was slightly altered. Daphnia inoculated with diet microbial inoculum has almost the same microbiome in both genotypes. The total number of neonates and body size were significantly reduced in Daphnia inoculated with diet microbial inoculum regardless of genotype compared to all other treatments. Overall, this study shows that the microbiome of Daphnia is flexible and varies with genotype and diet- and medium-associated microbes, but not every bacteria is beneficial to Daphnia, and only symbionts can increase Daphnia performance.







Similar content being viewed by others
Data Availability
Sequence data is deposited in the Sequence Read Database (SRA) and available online under BioProject PRJNA737109. All other data will be available when requested.
Code Availability
Not applicable.
References
Sommer F, Bäckhed F (2013) The gut microbiota-masters of host development and physiology. Nat Rev Microbiol 11:227–238. https://doi.org/10.1038/nrmicro2974
Adair KL, Douglas AE (2017) Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol 35:23–29. https://doi.org/10.1016/j.mib.2016.11.002
Peixoto RS, Harkins DM, Nelson KE (2021) Advances in microbiome research for animal health. Microb Ecol 9:289–311. https://doi.org/10.1146/annurev-animal-091020-075907
Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S (2012) Host-gut microbiota metabolic interactions. Science 336:1262–1267. https://doi.org/10.1126/science.1223813
Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ (2010) Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329:212–215. https://doi.org/10.1126/science.1188235
Koch H, Schmid-Hempel P (2011) Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci 108:19288–19292. https://doi.org/10.1073/pnas.1110474108
Erica VH, Jacobus CR, Nicole MG (2019) Diet–microbiome–disease: investigating diet’s influence on infectious disease resistance through alteration of the gut microbiome. PLoS Pathog. 15(10):e1007891. https://doi.org/10.1371/journal.ppat.1007891
Spor A, Koren O, Ley R (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290. https://doi.org/10.1038/nrmicro2540
Frankel-Bricker J, Song MJ, Benner MJ, Schaack S (2019) Variation in the microbiota associated with Daphnia magna across genotypes, populations, and temperature. Microb Ecol 79:731–742. https://doi.org/10.1007/s00248-019-01412-9
Jackrel SL, Schmidt KC, Cardinale BJ, Denef VJ (2020) Microbiomes reduce their host’s sensitivity to interspecific interactions. mBio 11:e02657-19. https://doi.org/10.1128/mbio.02657-19
Taipale SJ, Brett MT, Pulkkinen K, Kainz MJ (2012) The influence of bacteria-dominated diets on Daphnia magna somatic growth, reproduction, and lipid composition. FEMS Microbiol Ecol 82:50–62. https://doi.org/10.1111/j.1574-6941.2012.01406.x
Lyu K, Guan H, Wu C, Wang X, Wilson AE, Yang Z (2016) Maternal consumption of non-toxic Microcystis by Daphnia magna induces tolerance to toxic Microcystis in offspring. Freshw Biol 61:219–228. https://doi.org/10.1111/fwb.12695
Lyu K, Cao C, Li D, Akbar S, Yang Z (2021) The thermal regime modifies the response of aquatic keystone species Daphnia to microplastics: evidence from population fitness, accumulation, histopathological analysis and candidate gene expression. Sci Total Environ 783:147154. https://doi.org/10.1016/j.scitotenv.2021.147154
Huang J, Li YR, Zhou QM, Sun YF, Zhang L, Gu L, Lyu K, Huang Y, Chen YF, Yang Z (2020) Non-toxic and toxic Microcystis aeruginosa reduce the tolerance of Daphnia pulex to low calcium in different degrees: based on the changes in the key life-history traits. Chemosphere 248:226101. https://doi.org/10.1016/j.chemosphere.2020.126101
Yang Z, Xiang F, Minter EJA, Lü K, Chen Y, Montagnes DJS (2011) The interactive effects of microcystin and nitrite on life-history parameters of the cladoceran Daphnia obtusa. J Hazard Mater 190:113–118. https://doi.org/10.1016/j.jhazmat.2011.03.002
Akbar S, Du JJ, Jia Y, Tian XJ (2017) The importance of calcium in improving resistance of Daphnia to Microcystis. PLoS ONE 12:e0175881. https://doi.org/10.1371/journal.pone.0175881
Bovyn RA, McCauley E, LaMontagne JM (2018) Offspring size-number tradeoffs and food quality feedbacks impact population dynamics in a Daphnia-algae system. Oikos 127:1152–1162. https://doi.org/10.1111/oik.04788
Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Knight R, Caporaso JG, Svanbäck R (2014) Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol Lett 17:979–987. https://doi.org/10.1111/ele.12301
Derrien M, Veiga P (2017) Rethinking diet to aid human-microbe symbiosis. Trends Microbiol 25:100–112. https://doi.org/10.1016/j.tim.2016.09.011
Seymour JR, Amin SA, Raina JB, Stocker R (2017) Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol 2:17065. https://doi.org/10.1038/nmicrobiol.2017.65
Schmidt KC, Jackrel SL, Smith DJ, Dick GJ, Denef VJ (2020) Genotype and host microbiome alter competitive interactions between Microcystis aeruginosa and Chlorella sorokiniana. Harmful Algae 99:101939. https://doi.org/10.1016/j.hal.2020.101939
Jackrel SL, Yang JW, Schmidt KC, Denef VJ (2021) Host specificity of microbiome assembly and its fitness effects in phytoplankton. ISME J 15:774–788. https://doi.org/10.1038/s41396-020-00812-x
Johnson AJ, Zheng JJ, Kang JW, Saboe A, Knights D & Zivkovic AM (2020) A guide to diet-microbiome study design. Front Nutr 7. https://doi.org/10.3389/fnut.2020.00079
Zhang X, Ohtsuki H, Makino W, Kato Y, Watanabe H, Urabe J (2021) Variations in effects of ectosymbiotic microbes on the growth rates among different species and genotypes of Daphnia fed different algal diets. Ecol Res 36:303–312. https://doi.org/10.1111/1440-1703.12194
Sullam KE, Pichon S, Schaer TMM, Ebert D (2018) The combined effect of temperature and host clonal line on the microbiota of a planktonic crustacean. Microb Ecol 76:506–517. https://doi.org/10.1007/s00248-017-1126-4
Callens M, Watanabe H, Kato Y, Miura J, Decaestecker E (2018) Microbiota inoculum composition affects holobiont assembly and host growth in Daphnia. Microbiome 6:56. https://doi.org/10.1186/s40168-018-0444-1
Callens M, Macke E, Muylaert K, Bossier P, Lievens B, Waud M, Decaestecker E (2015) Food availability affects the strength of mutualistic host-microbiota interactions in Daphnia magna. ISME J 10:911–920. https://doi.org/10.1038/ismej.2015.166
Mushegian AA, Ebert D (2017) Presence of microbiota reverses the relative performance of Daphnia on two experimental diets. Zoology 125:29–31. https://doi.org/10.1016/j.zool.2017.07.007
Cooper RO, Cressler CE (2020) Characterization of key bacterial species in the Daphnia magna microbiota using shotgun metagenomics. Sci Rep 10:652. https://doi.org/10.1038/s41598-019-57367-x
Macke E, Callens M, De Meester L, Decaestecker E (2017) Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat Commu 8:1608. https://doi.org/10.1038/s41467-017-01714-x
Smith DJ, Tan JY, Powers MA, Lin XN, Davis TW, Dick GJ (2021) Individual Microcystis colonies harbour distinct bacterial communities that differ by Microcystis oligotype and with time. Environ Microbiol. https://doi.org/10.1111/1462-2920.15514
Cirri E, Pohnert G (2019) Algae-bacteria interactions that balance the planktonic microbiome. New Phytol 223:100–106. https://doi.org/10.1111/nph.15765
Akbar S, Gu L, Sun YF, Zhou QM, Zhang L, Lyu K, Yuan H, Yang Z (2020) Changes in the life history traits of Daphnia magna are associated with the gut microbiota composition shaped by diet and antibiotics. Sci Total Environ 705:135827. https://doi.org/10.1016/j.scitotenv.2019.135827
Akbar S, Huang J, Zhou QM, Gu L, Sun YF, Zhang L, Lyu K, Yang Z (2021) Elevated temperature and toxic Microcystis reduce Daphnia fitness and modulate gut microbiota. Environ Pollut 271:116409. https://doi.org/10.1016/j.envpol.2020.116409
Cooper RO, Vavra JM, Cressler CE (2021) Targeted manipulation of abundant and rare taxa in the Daphnia magna microbiota with antibiotics impacts host fitness differentially. mSystems 6:e00916-20. https://doi.org/10.1128/msystems.00916-20
Eckert EM, Anicic N, Fontaneto D (2021) Freshwater zooplankton microbiome composition is highly flexible and strongly influenced by the environment. Mol Ecol 30:1545–1558. https://doi.org/10.1111/mec.15815
Freese HM, Schink B (2011) Composition and stability of the microbial community inside the digestive tract of the aquatic crustacean Daphnia magna. Microb Ecol 62:882–894. https://doi.org/10.1007/s00248-011-9886-8
Eigemann F, Hilt S, Salka I, Grossart HP (2013) Bacterial community composition associated with freshwater algae: species specificity vs. dependency on environmental conditions and source community. FEMS Microbiol Ecol 83:650–663. https://doi.org/10.1111/1574-6941.12022
Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S (2017) The evolution of the host microbiome as an ecosystem on a leash. Nature 548:43–51. https://doi.org/10.1038/nature23292
Mushegian AA, Walser JC, Sullam KE, Ebert D (2018) The microbiota of diapause: how host-microbe associations are formed after dormancy in an aquatic crustacean. J Anim Ecol 87:400–413. https://doi.org/10.1111/1365-2656.12709
Peerakietkhajorn S, Tsukada K, Kato Y, Matsuura T, Watanabe H (2015) Symbiotic bacteria contribute to increasing the population size of a freshwater crustacean, Daphnia magna. Environ Microbiol Rep 7:364–372. https://doi.org/10.1111/1758-2229.12260
Marinho MC, Lage OM, Catita J, Antunes SC (2018) Adequacy of planctomycetes as supplementary food source for Daphnia magna. Antonie Van Leeuwenhoek 111:825–840. https://doi.org/10.1007/s10482-017-0997-1
Sison-Mangus MP, Mushegian AA, Ebert D (2015) Water fleas require microbiota for survival, growth and reproduction. ISME J 9:59–67. https://doi.org/10.1038/ismej.2014.116
Ruuskanen MO, Sommeria-Klein G, Havulinna AS, Niiranen TJ, Lahti L (2021) Modelling spatial patterns in host-associated microbial communities. Environ Microbiol 23:2374–2388. https://doi.org/10.1111/1462-2920.15462
Grainger TN, Letten AD, Gilbert B, Fukami T (2019) Applying modern coexistence theory to priority effects. Proc Natl Acad Sci 116:6205–6210. https://doi.org/10.1073/pnas.1803122116
Li WZ, Nelson KE (2021) Microbial species that initially colonize the human gut at birth or in early childhood can stay in human body for lifetime. Microb Ecol. https://doi.org/10.1007/s00248-020-01636-0
Callens M, De Meester L, Muylaert K, Mukherjee S, Decaestecker E (2020) The bacterioplankton community composition and a host genotype dependent occurrence of taxa shape the Daphnia magna gut bacterial community. FEMS Microbiol Ecol 96. https://doi.org/10.1093/femsec/fiaa128
Houwenhuyse S, Stoks R, Mukherjee S, Decaestecker E (2021) Locally adapted gut microbiomes host stress tolerance. ISME J. https://doi.org/10.1038/s41396-021-00940-y
Funding
This work was supported by the National Natural Science Foundation of China (31730105 and 31800385), China Postdoctoral Science Foundation (2020M681658), and the Priority Academic Program Development of Jiangsu Higher Education Institutions of China.
Author information
Authors and Affiliations
Contributions
S.A and Z.Y conceptualize and designed the present study. S.A, Q.L, X.L, L.G, and Z.D performed experiments. S.A, J.H, and Q.Z analyzed the data. S.A wrote the manuscript. S.A and Z.Y revised and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
The authors declare their consent to participate in the present work.
Consent for Publication
The authors declare their consent for the publication of the present work.
Competing Interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Akbar, S., Li, X., Ding, Z. et al. Disentangling Diet- and Medium-Associated Microbes in Shaping Daphnia Gut Microbiome. Microb Ecol 84, 911–921 (2022). https://doi.org/10.1007/s00248-021-01900-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01900-x