Abstract
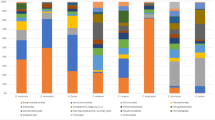
Animals host a wide diversity of symbiotic microorganisms that contribute important functions to host health, and our knowledge of what drives variation in the composition of these complex communities continues to grow. Microbiome studies at larger spatial scales present opportunities to evaluate the contribution of large-scale factors to variation in the microbiome. We conducted a large-scale field study to assess variation in the bacterial symbiont communities on adult frog skin (Pseudacris crucifer), characterized using 16S rRNA gene amplicon sequencing. We found that skin bacterial communities on frogs were less diverse than, and structurally distinct from, the surrounding habitat. Frog skin was typically dominated by one of two bacterial OTUs: at western sites, a Proteobacteria dominated the community, whereas eastern sites were dominated by an Actinobacteria. Using a metacommunity framework, we then sought to identify factors explaining small- and large-scale variation in community structure—that is, among hosts within a pond, and among ponds spanning the study transect. We focused on the presence of a fungal skin pathogen, Batrachochytrium dendrobatidis (Bd) as one potential driver of variation. We found no direct link between skin bacterial community structure and Bd infection status of individual frog hosts. Differences in pond-level community structure, however, were explained by Bd infection prevalence. Importantly, Bd infection prevalence itself was correlated with numerous other environmental factors; thus, skin bacterial diversity may be influenced by a complex suite of extrinsic factors. Our findings indicate that large-scale factors and processes merit consideration when seeking to understand microbiome diversity.



Similar content being viewed by others
References
Adair KL, Douglas AE (2017) Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol 35:23–29
Griffiths SM, Harrison XA, Weldon C, Wood MD, Pretorius A, Hopkins K, Fox G, Preziosi RF, Antwis RE (2018) Genetic variability and ontogeny predict microbiome structure in a disease-challenged montane amphibian. ISME J 12:2506–2517
SanMiguel A, Grice EA (2015) Interactions between host factors and the skin microbiome. Cell Mol Life Sci 72:1499–1515
Ricklefs RE (1987) Community diversity: relative roles of local and regional processes. Science 235:167–171
Lindström ES, Langenheder S (2012) Local and regional factors influencing bacterial community assembly. Environ Microbiol Rep 4:1–9
Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356
Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA (2012) The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262
Fierer N, Ferrenberg S, Flores GE, González A, Kueneman J, Legg T, Lynch RC, McDonald D, Mihaljevic JR, O’Neill SP, Rhodes ME, Song SJ, Walters WA (2012) From animalcules to an ecosystem: application of ecological concepts to the human microbiome. Annu Rev Ecol Evol Syst 43:137–155
Mihaljevic JR (2012) Linking metacommunity theory and symbiont evolutionary ecology. Trends Ecol Evol 27:323–329
Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, Holt RD, Shurin JB, Law R, Tilman D, Loreau M, Gonzalez A (2004) The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett 7:601–613
Holyoak M, Leibold MA, Holt RD (eds.) (2005) Metacommunities: spatial dynamics and ecological communities. University of Chicago Press
Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, Schmidt TM (2015) Application of a neutral community model to assess structuring of the human lung microbiome. MBio 6:e02284–e02214
Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ (2016) Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10:655–664
Bletz MC, Loudon AH, Becker MH, Bell SC, Woodhams DC, Minbiole KPC, Harris RN (2013) Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol Lett 16:807–820
Rebollar EA, Antwis RE, Becker MH, Belden LK, Bletz MC, Brucker RM, Harrison XA, Hughey MC, Kueneman JG, Loudon AH, McKenzie V (2016) Using “omics” and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front Microbiol 7:68
Woodhams DC, Bletz M, Kueneman J, McKenzie V (2016) Managing amphibian disease with skin microbiota. Trends Microbiol 24:161–164
Longo AV, Savage AE, Hewson I, Zamudio KR (2015) Seasonal and ontogenetic variation of skin microbial communities and relationships to natural disease dynamics in declining amphibians. R Soc Open Sci 2:140377
Krynak KL, Burke DJ, Benard MF (2016) Landscape and water characteristics correlate with immune defense traits across Blanchard’s cricket frog (Acris blanchardi) populations. Biol Conserv 193:153–167
Becker CG, Longo AV, Haddad CF, Zamudio KR (2017) Land cover and forest connectivity alter the interactions among host, pathogen and skin microbiome. Proc R Soc B 284:20170582
Hughey MC, Pena JA, Reyes R, Medina D, Belden LK, Burrowes PA (2017) Skin bacterial microbiome of a generalist Puerto Rican frog varies along elevation and land use gradients. PeerJ 5:e3688
Muletz Wolz CR, Yarwood SA, Campbell Grant EH, Fleischer RC, Lips KR (2018) Effects of host species and environment on the skin microbiome of Plethodontid salamanders. Anim Ecol 87:341–353
Jani AJ, Briggs CJ (2014) The pathogen Batrachochytrium dendrobatidis disturbs the frog skin microbiome during a natural epidemic and experimental infection. 2014. Proc Natl Acad Sci 111:E5049–E5058
Belden LK, Hughey MC, Rebollar EA, Umile TP, Loftus SC, Burzynski EA, Minbiole KPC, House LL, Jensen RV, Becker MH, Walke JB, Medina D, Ibáñez R, Harris RN (2015) Panamanian frog species host unique skin bacterial communities. Front Microbiol 6:1171
Walke JB, Becker MH, Loftus SC, House LL, Teotonio TL, Minbiole KP, Belden LK (2015) Community structure and function of amphibian skin microbes: an experiment with bullfrogs exposed to a chytrid fungus. PLoS One 10:e0139848
Rebollar EA, Hughey MC, Medina D, Harris RN, Ibáñez R, Belden LK (2016) Skin bacterial diversity of Panamanian frogs is associated with host susceptibility and presence of Batrachochytrium dendrobatidis. ISME J 10:1682–1695
Jani AJ, Knapp RA, Briggs CJ (2017) Epidemic and endemic pathogen dynamics correspond to distinct host population microbiomes at a landscape scale. Proc R Soc B 284:20170944
Longo AV, Zamudio KR (2017) Environmental fluctuations and host skin bacteria shift survival advantage between frogs and their fungal pathogen. ISME J 11:349–361
Longo AV, Zamudio KR (2017) Temperature variation, bacterial diversity and fungal infection dynamics in the amphibian skin. Mol Ecol 26:4787–4797
Bates KA, Clare FC, O’Hanlon S, Bosch J, Brookes L, Hopkins K, McLaughlin EJ, Daniel O, Garner TW, Fisher MC, Harrison XA (2018) Amphibian chytridiomycosis outbreak dynamics are linked with host skin bacterial community structure. Nat Commun 9:693
Kilpatrick AM, Briggs CJ, Daszak P (2010) The ecology and impact of chytridiomycosis: an emerging disease of amphibians. Trends Ecol Evol 25:109–118
Lips KR (2016) Overview of chytrid emergence and impacts on amphibians. Philos Trans R Soc B 371:20150465
Voyles J, Rosenblum EB, Berger L (2011) Interactions between Batrachochytrium dendrobatidis and its amphibian hosts: a review of pathogenesis and immunity. Microbes Infect 13:25–32
Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC, Lam BA, Woodhams DC, Briggs CJ, Vredenburg VT, Minbiole KPC (2009) Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J 3:818–824
Hughey MC, Becker MH, Walke JB, Swartwout MC, Belden LK (2014) Batrachochytrium dendrobatidis in Virginia amphibians: within and among site variation in infection. Herpetol Rev 45:428–438
Gahl MK, Longcore JE, Houlahan JE (2012) Varying responses of northeastern north American amphibians to the chytrid pathogen Batrachochytrium dendrobatidis. Conserv Biol 26:135–141
Hyatt AD, Boyle DG, Olsen V, Boyle DB, Berger L, Obendorf D, Dalton A, Kriger K, Hero M, Hines H, Phillott R, Campbell R, Marantelli G, Gleason F, Colling A (2007) Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis Aquat Org 73:175–192
McKenzie VJ, Bowers RM, Fierer N, Knight R, Lauber CL (2012) Co-habiting amphibian species harbor unique skin bacterial communities in wild populations. ISME J 6:588–596
Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV, Belden LK (2014) Amphibian skin may select for rare environmental microbes. ISME J 8:2207–2217
Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD (2004) Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Org 60:141–148
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Meisel JS, Hannigan GD, Tyldsley AS, SanMiguel AJ, Hodkinson BP, Zheng Q, Grice EA (2016) Skin microbiome surveys are strongly influenced by experimental design. J Invest Dermatol 136:947–956
Aronesty E. (2011) Ea-utils: “command-line tools for processing biological sequencing data”. http://code.google.com/p/ea-utils
Caporaso JG, Kuczynski J, Stombaugh JJ, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
R Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna http://www.R-project.org
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2015) Vegan: community ecology package. http://CRAN.R-project.org/package=vegan
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Aust Ecol 26:32–46
Whittaker RH (1972) Evolution and measurement of species diversity. Taxon 21:213–251
Fournier DA, Skaug HJ, Ancheta J, Ianelli J, Magnusson A, Maunder M et al (2012) AD model builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models. Optim Methods Softw 27:233–249
Skaug H, Fournier D, Bolker B, Magnusson A, Nielsen A (2014) Generalized linear mixed models using AD model builder. R package version 0.8.0
Legendre P, Legendre L (2012) Numerical ecology. 3rd English edition. Elsevier
Tuomisto H, Ruokolainen K (2006) Analyzing or explaining beta diversity? Understanding the targets of different methods of analysis. Ecology 87:2697–2708
Paliy O, Shankar V (2016) Application of multivariate statistical techniques in microbial ecology. Mol Ecol 25:1032–1057
Legendre P, Gallagher ED (2001) Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280
Campbell BJ, Yu L, Heidelberg JF, Kirchman DL (2011) Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci U S A 108:12776–12781
Walke JB, Becker MH, Hughey MC, Swartwout MC, Jensen RV, Belden LK (2017) Dominance-function relationships in the amphibian skin microbiome. Environ Microbiol 19:3387–3397
Austin JD, Lougheed SC, Neidrauer L, Chek AA, Boag PT (2002) Cryptic lineages in a small frog: the post-glacial history of the spring peeper, Pseudacris crucifer (Anura: Hylidae). Mol Phylogenet Evol 25:316–329
Christian N, Whitaker BK, Clay K (2015) Microbiomes: unifying animal and plant systems through the lens of community ecology theory. Front Microbiol 6:869
Burns AR, Miller E, Agarwal M, Rolig AS, Milligan-Myhre K, Seredick S, Guillemin K, Bohannan BJ (2017) Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc Natl Acad Sci 114:11181–11186
Fitzpatrick BM, Allison AL (2014) Similarity and differentiation between bacteria associated with skin of salamanders (Plethodon jordani) and free-living assemblages. FEMS Microbiol Ecol 88:482–494
Banning JL, Weddle AL, Wahl GW, Simon MA, Lauer A, Walters RL, Harris RN (2008) Antifungal skin bacteria, embryonic survival, and communal nesting in four-toed salamanders, Hemidactylium scutatum. Oecologia 156:423–429
Walke JB, Harris RN, Reinert LK, Rollins-Smith LA, Woodhams DC (2011) Social immunity in amphibians: evidence for vertical transmission of innate defenses. Biotropica 43:396–400
Hughey MC, Delia J, Belden LK (2017) Diversity and stability of egg-bacterial assemblages: the role of paternal care in the glassfrog Hyalinobatrachium colymbiphyllum. Biotropica 49:792–802
Acknowledgments
We thank P. Shirk, J. Vonesh, J. Wyderko, and S. Zemmer for assistance in the field; P. Sattler at Liberty University, G. Eaton at Claytor Nature Study Center, and J. Vonesh at Virginia Commonwealth University for recommending sites in Lynchburg, Bedford, and Richmond, respectively; J. Touchon for R code consultations; C. Herbold for stats advice; and Z. Herbert at the Dana Farber Cancer Institute Molecular Biology Core Facility for Illumina MiSeq amplicon sequencing. We are also thankful for the comments of anonymous reviewers that improved the manuscript.
Funding
This work was supported by the Morris Animal Foundation [D10ZO-028] and the National Science Foundation [DEB-1136640].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This research was conducted with approval from Virginia Tech’s Institutional Animal Care and Use Committee (protocol 10-029-BIOL) and with permission from the Virginia Department of Game and Inland Fisheries (permit 44303).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(R 3 kb)
Rights and permissions
About this article
Cite this article
Hughey, M.C., Sokol, E.R., Walke, J.B. et al. Ecological Correlates of Large-Scale Turnover in the Dominant Members of Pseudacris crucifer Skin Bacterial Communities. Microb Ecol 78, 832–842 (2019). https://doi.org/10.1007/s00248-019-01372-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01372-0