Abstract
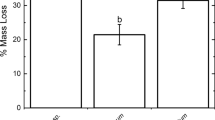
It has been suggested that food storage inside the nest may offer termites with a nutritional provision during low resource availability. Additionally, feces employed as construction material provide an excellent environment for colonization by microorganisms and, together with the storage of plant material inside the nest, could thus provide some advantage to the termites in terms of lignocellulose decomposition. Here, we conducted for the first time a comprehensive study of the microbial communities associated to a termite exhibiting food storage behavior using Illumina sequencing of the 16S and (ITS2) regions of rRNA genes, together with enzymatic assays and data collected in the field. Cornitermes cumulans (Syntermitinae) stored grass litter in nodules made from feces and saliva located in the nest core. The amount of nodules increased with nest size and isolation, and interestingly, the soluble fraction of extracts from nodules showed a higher activity against hemicellulosic substrates compared to termite guts. Actinobacteria and Sordariales dominated microbial communities of food nodules and nest walls, whereas Spirochetes and Pleosporales dominated gut samples of C. cumulans. Within Syntermitinae, however, gut bacterial assemblages were dissimilar. On the other hand, there is a remarkable convergence of the bacterial community structure of Termitidae nests. Our results suggest that the role of nodules could be related to food storage; however, the higher xylanolytic activity in the nodules and their associated microbiota could also provide C. cumulans with an external source of predigested polysaccharides, which might be advantageous in comparison with litter-feeding termites that do not display food storage behavior.




Similar content being viewed by others
References
Brune A (2014) Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. https://doi.org/10.1038/nrmicro3182
Neupane A, Maynard DS, Bradford MA (2015) Consistent effects of eastern subterranean termites (Reticulitermes flavipes) on properties of a temperate forest soil. Soil Biol Biochem 91:84–91. https://doi.org/10.1016/j.soilbio.2015.08.025
Siebers N, Martius C, Eckhardt K-U et al (2015) Origin and alteration of organic matter in termite mounds from different feeding guilds of the Amazon rainforests. PLoS One 10:e0123790. https://doi.org/10.1371/journal.pone.0123790
Breznak JA, Brune A (1994) Role of the microorganisms in the digestion of lignocellulose by termites. Annu Rev Entomol 39:453–487
Dietrich C, Köhler T, Brune A (2014) The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol. https://doi.org/10.1128/AEM.04206-13
Nalepa CA, Bignell DE, Bandi C (2001) Detritivory, coprophagy, and the evolution of digestive mutualisms in Dictyoptera. Insect Soc 48:194–201. https://doi.org/10.1007/PL00001767
Engel MS, Grimaldi DA, Krishna K (2009) Termites (Isoptera): their phylogeny, classification, and rise to ecological dominance. Am Mus Novit 3650:1–27. https://doi.org/10.1206/651.1
Mikaelyan A, Strassert JFH, Tokuda G, Brune A (2014) The fibre-associated cellulolytic bacterial community in the hindgut of wood-feeding higher termites (Nasutitermes spp.). Environ Microbiol 16:2711–2722. https://doi.org/10.1111/1462-2920.12425
Donovan SE, Eggleton P, Bignell DE (2001) Gut content analysis and a new feeding group classification of termites. Ecol Entomol 26:356–366. https://doi.org/10.1046/j.1365-2311.2001.00342.x
Rahman NA, Parks DH, Willner DL et al (2015) A molecular survey of Australian and North American termite genera indicates that vertical inheritance is the primary force shaping termite gut microbiomes. Microbiome 3:5. https://doi.org/10.1186/s40168-015-0067-8
Mikaelyan A, Dietrich C, Köhler T et al (2015) Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol Ecol 24:5284–5295. https://doi.org/10.1111/mec.13376
Hongoh Y, Ekpornprasit L, Inoue T et al (2006) Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol Ecol 15:505–516. https://doi.org/10.1111/j.1365-294X.2005.02795.x
Rocha MM, Morales-Corrêa e Castro AC, Cuezzo C et al (2017) Phylogenetic reconstruction of Syntermitinae (Isoptera, Termitidae) based on morphological and molecular data. PLoS One 12:e0174366. https://doi.org/10.1371/journal.pone.0174366
Emerson AE (1952) The neotropical genera Procornitermes and Cornitermes (Isoptera, Termitidae). Bull Am Mus Nat Hist 99:479–539
Schmidt AM, Jacklyn P, Korb J (2014) “Magnetic” termite mounds: is their unique shape an adaptation to facilitate gas exchange and improve food storage? Insect Soc 61:41–49. https://doi.org/10.1007/s00040-013-0322-6
Lima JT, Costa-Leonardo AM (2007) Recursos alimentares explorados pelos cupins (Insecta: Isoptera). Biota Neotrop 7:243–250. https://doi.org/10.1590/S1676-06032007000200027
Holt J (1998) Microbial activity in the mounds of some Australian termites. Appl Soil Ecol 9:183–187. https://doi.org/10.1016/S0929-1393(98)00073-0
Redford KH (1984) The termitaria of Cornitermes cumulans (Isoptera, Termitidae) and their role in determining a potential keystone species. Biotropica 16:112–119. https://doi.org/10.2307/2387842
Darlington JPEC, Dransfield RD (1987) Size relationships in nest populations and mound parameters in the termite Macrotermes michaelseni in Kenya. Insect Soc 34:165–180. https://doi.org/10.1007/BF02224082
Acioli ANS, Constantino R (2015) A taxonomic revision of the neotropical termite genus Ruptitermes (Isoptera, Termitidae, Apicotermitinae). Zootaxa 4032:451–492. https://doi.org/10.11646/zootaxa.4032.5.1
Coles De Negret HR, Redford KH (1982) The biology of nine termite species (Isoptera: Termitidae) from the Cerrado of Central Brazil. Psyche (Stuttg) 89:81–106
AOAC (2006) Official Methods of Analysis of AOAC INTERNATIONAL. 18th ed. AOAC INTERNATIONAL, Gaithersburg
Van Soest PJ (1963) Use of detergents in the analysis of fibrous feeds. II. A rapid method for the determination of fiber and lignin. J AOAC 46:829–835
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Franco Cairo JPL, Leonardo FC, Alvarez TM et al (2011) Functional characterization and target discovery of glycoside hydrolases from the digestome of the lower termite Coptotermes gestroi. Biotechnol Biofuels 4:50. https://doi.org/10.1186/1754-6834-4-50
Yu Z, Morrison M (2004) Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 36:808–812. https://doi.org/10.2144/3605A0808
Caporaso JG, Lauber CL, Walters WA et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. https://doi.org/10.1073/pnas.1000080107
White TJ, Bruns S, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–322
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Bengtsson-Palme J, Ryberg M, Hartmann M et al (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. https://doi.org/10.1111/2041-210X.12073
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Mikaelyan A, Köhler T, Lampert N et al (2015) Classifying the bacterial gut microbiota of termites and cockroaches: a curated phylogenetic reference database (DictDb). Syst Appl Microbiol 38:472–482. https://doi.org/10.1016/j.syapm.2015.07.004
Deshpande V, Wang Q, Greenfield P et al (2016) Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia 108:1–5. https://doi.org/10.3852/14-293
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. https://doi.org/10.1371/journal.pone.0061217
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria ISBN 3-900051-07-0, URL http://www.R-project.org/55:275–286
Pinheiro J, Bates D, DebRoy S, Sarkar D (2016) nlme: linear and nonlinear mixed effects models. R Packag version R package, pp 1–86
Lenth RV (2016) Least-squares means: the R package lsmeans. J Stat Softw 69:1–33. https://doi.org/10.18637/jss.v069.i01
Graves S, Piepho H-P, Selzer L Sundar DR, et al (2015) Package “multcompView” visualizations of paired comparisons. R Packag. https://CRAN.R-project.org/package=multcompView
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. https://doi.org/10.2307/2346101
Wickham H (2009) ggplot2 elegant graphics for data analysis. Media. https://doi.org/10.1007/978-0-387-98141-3
Neuwirth E (2014) RColorBrewer: ColorBrewer palettes. R Packag version, pp 11–2. https://cran.R-project.org/package=RColorBrewer
Oksanen J, Blanchet FG, Kindt R, et al (2013) Package “vegan.” R Packag ver 20–8 254. https://doi.org/10.4135/9781412971874.n145
Anderson MJ, Ellingsen KE, McArdle BH (2006) Multivariate dispersion as a measure of beta diversity. Ecol Lett 9:683–693. https://doi.org/10.1111/j.1461-0248.2006.00926.x
De Caceres M, Jansen F(2011) Package “indicspecies.” probability.ca, pp 1–16
Korb J (2003) The shape of compass termite mounds and its biological significance. Insect Soc 50:218–221
Thorne BL, Collins MS, Bjorndal KA (1996) Architecture and nutrient analysis of arboreal carton nests of two neotropical Nasutitermes species (Isoptera: Termitidae), with notes on embedded nodules. Florida Entomol 79:27–37
Franco Cairo JPL, Carazzolle MF, Leonardo FC et al (2016) Expanding the knowledge on lignocellulolytic and redox enzymes of worker and soldier castes from the lower termite Coptotermes gestroi. Front Microbiol 7:1518. https://doi.org/10.3389/fmicb.2016.01518
He S, Ivanova N, Kirton E et al (2013) Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One 8:e61126. https://doi.org/10.1371/journal.pone.0061126
Brauman A, Bignell DE, Tayasu I (2000) Termites: evolution, sociality, symbioses, ecology. In: Abe T, Bignell DE, Higashi M (eds) Termites: evolution, sociality, symbioses, ecology. Kluwer Academic, Dordrecht, pp 233–259
Hu J, Arantes V, Saddler JN (2011) The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: is it an additive or synergistic effect? Biotechnol Biofuels 4:36. https://doi.org/10.1186/1754-6834-4-36
Tokuda G, Watanabe H (2007) Hidden cellulases in termites: revision of an old hypothesis. Biol Lett 3:336–339. https://doi.org/10.1098/rsbl.2007.0073
Eriksson T, Börjesson J, Tjerneld F (2002) Mechanism of surfactant effect in enzymatic hydrolysis of lignocellulose. Enzym Microb Technol 31:353–364. https://doi.org/10.1016/S0141-0229(02)00134-5
Alkasrawi M, Eriksson T, Börjesson J et al (2003) The effect of Tween-20 on simultaneous saccharification and fermentation of softwood to ethanol. Enzym Microb Technol 33:71–78. https://doi.org/10.1016/S0141-0229(03)00087-5
Fall S, Hamelin J, Ndiaye F et al (2007) Differences between bacterial communities in the gut of a soil-feeding termite (Cubitermes niokoloensis) and its mounds. Appl Environ Microbiol 73:5199–5208. https://doi.org/10.1128/AEM.02616-06
Cosarinsky MI (2011) The nest growth of the neotropical mound-building termite, Cornitermes cumulans: a micromorphological analysis. J Insect Sci 11:122. https://doi.org/10.1673/031.011.12201
Visser AA, Nobre T, Currie CR et al (2012) Exploring the potential for actinobacteria as defensive symbionts in fungus-growing termites. Microb Ecol 63:975–985. https://doi.org/10.1007/s00248-011-9987-4
Kuhnigk T, Branke J, Krekeler D et al (1996) A feasible role of sulfate-reducing bacteria in the termite gut. Syst Appl Microbiol 19:139–149. https://doi.org/10.1016/S0723-2020(96)80039-7
Mohana S, Shah A, Divecha J, Madamwar D (2008) Xylanase production by Burkholderia sp. DMAX strain under solid state fermentation using distillery spent wash. Bioresour Technol 99:7553–7564. https://doi.org/10.1016/j.biortech.2008.02.009
Costa PS, Oliveira PL, Chartone-Souza E, Nascimento AMA (2013) Phylogenetic diversity of prokaryotes associated with the mandibulate nasute termite Cornitermes cumulans and its mound. Biol Fertil Soils 49:567–574. https://doi.org/10.1007/s00374-012-0742-x
Warnecke F, Luginbühl P, Ivanova N et al (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565. https://doi.org/10.1038/nature06269
Mikaelyan A, Meuser K, Brune A (2017) Microenvironmental heterogeneity of gut compartments drives bacterial community structure in wood- and humus-feeding higher termites. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiw210
Otani S, Hansen LH, Sørensen SJ, Poulsen M (2016) Bacterial communities in termite fungus combs are comprised of consistent gut deposits and contributions from the environment. Microb Ecol 71:207–220. https://doi.org/10.1007/s00248-015-0692-6
Stackebrandt E (2014) The family Lachnospiraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes: firmicutes and tenericutes. Springer, Berlin, pp 197–201
Constantino R (2015) Cupins do Cerrado. Technical Books, Rio de Janeiro
Machida M, Kitade O, Miura T, Matsumoto T (2001) Nitrogen recycling through proctodeal trophallaxis in the Japanese damp-wood termite Hodotermopsis japonica (Isoptera, Termopsidae). Insect Soc 48:52–56. https://doi.org/10.1007/PL00001745
Poulsen M, Hu H, Li C et al (2014) Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc Natl Acad Sci U S A 111:14500–14505. https://doi.org/10.1073/pnas.1319718111
Scully ED, Geib SM, Hoover K et al (2013) Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle. PLoS One 8:e73827. https://doi.org/10.1371/journal.pone.0073827
Ni J, Tokuda G (2013) Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol Adv 31:838–850. https://doi.org/10.1016/j.biotechadv.2013.04.005
Acknowledgements
We thank Johana Rincones, Tiago Carrijo, and two anonymous reviewers for their comments on the manuscript. We would like to acknowledge the CTBE-NGS facility at the Brazilian Bioethanol Science and Technology Laboratory for provision of time and technical support.
Funding
This study was supported by the São Paulo Research Foundation (FAPESP), grant # 2015/21497-6 (A. A.), and the Minas Gerais State Agency for Research and Development – FAPEMIG, grant # CRA-APQ-00878-12 (A. A. and V. X. S.). L. M. was supported by a master degree grant from the Coordination for the Improvement of Higher Education Personnel (CAPES).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Menezes, L., Alvarez, T.M., Persinoti, G.F. et al. Food Storage by the Savanna Termite Cornitermes cumulans (Syntermitinae): a Strategy to Improve Hemicellulose Digestibility?. Microb Ecol 76, 492–505 (2018). https://doi.org/10.1007/s00248-017-1128-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-1128-2