Abstract
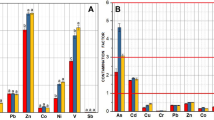
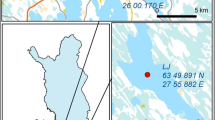
The deposition of mine tailings generated from 125 years of sulfidic ore mining resulted in the enrichment of Coeur d'Alene River (CdAR) sediments with significant amounts of toxic heavy metals. A review of literature suggests that microbial populations play a pivotal role in the biogeochemical cycling of elements in such mining-impacted sedimentary environments. To assess the indigenous microbial communities associated with metal-enriched sediments of the CdAR, high-density 16S microarray (PhyloChip) and clone libraries specific to bacteria (16S rRNA), ammonia oxidizers (amoA), and methanogens (mcrA) were analyzed. PhyloChip analysis provided a comprehensive assessment of bacterial populations and detected the largest number of phylotypes in Proteobacteria followed by Firmicutes and Actinobacteria. Furthermore, PhyloChip and clone libraries displayed considerable metabolic diversity in indigenous microbial populations by capturing several chemolithotrophic groups such as ammonia oxidizers, iron-reducers and -oxidizers, methanogens, and sulfate-reducers in the CdAR sediments. Twenty-two phylotypes detected on PhyloChip could not be classified even at phylum level thus suggesting the presence of novel microbial populations in the CdAR sediments. Clone libraries demonstrated very limited diversity of ammonia oxidizers and methanogens in the CdAR sediments as evidenced by the fact that only Nitrosospira- and Methanosarcina-related phylotypes were retrieved in amoA and mcrA clone libraries, respectively.





Similar content being viewed by others
References
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Baker BJ, Banfield JF (2003) Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44:139–152
Balistrieri LS, Box SE, Tonkin JW (2003) Modeling precipitation and sorption of elements during mixing of river water and pore water in the Coeur d'Alene river basin. Environ Sci Technol 37:4694–4701
Besser JM, Brumbaugh WG, Ivey CD, Ingersoll CG, Moran PW (2008) Biological and chemical characterization of metal bioavailability in sediments from Lake Roosevelt, Columbia River, Washington, USA. Arch Environ Contam Toxicol 54:557–570
Bouskill NJ, Barker-Finkel J, Galloway TS, Handy RD, Ford TE (2010) Temporal bacterial diversity associated with metal-contaminated river sediments. Ecotoxicology 19:317–328
Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL (2007) Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA 104:299–304
Chhatwal I (2008) Copper and zinc induced growth inhibition to Coeur d'Alene microbial isolates. Thesis South Dakota School of Mines and Technology, Rapid City, SD USA
Cummings DE, Caccavo F, Stefan S, Rosenzweiz RF (1999) Ferribacterium limneticum, gen. nov., sp. nov., an Fe(III)-reducing microorganism isolated from mining-impacted freshwater lake sediments. Arch Microbiol 171:183–188
Cummings DE, Caccavo F Jr, Fendorf S, Rosenzweig RF (1999) Arsenic mobilization by the dissimilatory Fe(III)-reducing bacterium Shewanella alga BrY. Environ Sci Technol 33:723–729
Cummings DE, March AW, Bostick B, Spring S, Caccavo F Jr, Fendorf S, Rosenzweig RF (2000) Evidence for microbial Fe(III) reduction in anoxic, mining-impacted Lake sediments (Lake Coeur d'Alene, Idaho). Appl Environ Microbiol 66:154–162
Cummings DE, Snoeyenbos-West OL, Newby DT, Niggemyer AM, Lovley DR, Achenbach LA, Rosenzweig RF (2003) Diversity of Geobacteraceae species inhabiting metal-polluted freshwater Lake sediments ascertained by 16S rDNA analyses. Microb Ecol 46:257–269
DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL (2007) High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol 53:371–383
Felsenstein J (1989) PHYLIP-Phylogeny inference package (version 3.2). Cladistics 5:164–166
Feris K, Ramsey P, Frazar C, Moore JN, Gannon JE, Holben WE (2003) Differences in hyporheic-zone microbial community structure along a heavy-metal contamination gradient. Appl Environ Microbiol 69:5563–5573
Feris KP, Ramsey PW, Frazar C, Rillig M, Moore JN, Gannon JE, Holben WE (2004) Seasonal dynamics of shallow-hyporheic-zone microbial community structure along a heavy-metal contamination gradient. Appl Environ Microbiol 70:2323–2331
Field EK, D’Imperio S, Miller AR, VanEngelen MR, Gerlach R, Lee BD, Apel WA, Peyton BM (2010) Application of molecular techniques to elucidate the influence of cellulosic waste on bacterial community structure at a simulated low level radioactive waste Site. Appl Environ Microbiol 76:3106–3115
Fortin D, Roy M, Rioux JP, Thibault PJ (2000) Occurrence of sulfate-reducing bacteria under a wide range of physico-chemical conditions in Au and Cu-Zn mine tailings. FEMS Microbiol Ecol 33:197–208
Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JL, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM (2008) Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603
Gillan DC, Danis B, Pernet P, Joly G, Dubois P (2005) Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment. Appl Environ Microbiol 71:679–690
Hallberg KB, Coupland K, Kimura S, Johnson DB (2006) Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl Environ Microbiol 72:2022–2030
Hanbo Z, Changqun D, Qiyong S, Weimin R, Tao S, Lizhong C, Zhiwei Z, Bin H (2004) Genetic and physiological diversity of phylogenetically and geographically distinct groups of Arthrobacter isolated from lead–zinc mine tailings. FEMS Microbiol Ecol 49:333–341
Hao C, Wang L, Gao Y, Zhang L, Dong H (2010) Microbial diversity in acid mine drainage of Xiang Mountain sulfide mine, Anhui Province, China. Extremophiles 14:465–474
Harrington JM, Fendorf SE, Rosenzweig RF (1998) Biotic generation of Arsenic(III) in metal(loid)-contaminated freshwater lake sediments. Environ Sci Technol 32:2425–2430
Harrington JM, Laforce MJ, Rember WC, Fendorf SE, Rosenzweig RF (1998) Phase associations and mobilization of iron and trace elements in Coeur d'Alene Lake, Idaho. Environ Sci Technol 32:650–656
Hemme CL, Deng Y, Gentry TJ, Fields MW, Wu L, Barua S, Barry K, Tringe SG, Watson DB, He Z, Hazen TC, Tiedje JM, Rubin EM, Zhou J (2010) Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J 4:660–672
Hernández-Eugenio G, Fardeau ML, Cayol JL, Patel BK, Thomas P, Macarie H, Garcia JL, Ollivier B (2002) Clostridium thiosulfatireducens sp. nov., a proteolytic, thiosulfate- and sulfur-reducing bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int J Syst Evol Microbiol 52:1461–1468
Horowitz AJ, Elrick KA (1995) Effect of mining and related activities on the sediment trace element geochemistry of Lake Coeur d'Alene, Idaho, USA Part II: subsurface sediments. Hydrol Process 9:35–54
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Kim BK, de Macario EC, Nölling J, Daniels L (1996) Isolation and characterization of a copper-resistant methanogen from a copper-mining soil sample. Appl Environ Microbiol 62:2629–2635
Konstantinidis KT, Isaacs N, Fett J, Simpson S, Long DT, Marsh TL (2003) Microbial diversity and resistance to copper in metal-contaminated lake sediment. Microb Ecol 45:191–202
Kumar S, Tamura K, Nei M (1993) MEGA: molecular evolutionary genetics analysis. Pennsylvania State University, University Park, PA
Moberly JG (2006) Biogeochemical cycling of toxic metals in Lake Coeur d’ Alene sediments. Thesis Washington State University, Pullman USA
Moberly JG, Borch T, Sani RK, Spycher NF, Sengor SS, Ginn TR, Peyton BM (2009) Heavy metal-mineral associations in Coeur d'Alene River sediments: a synchrotron-based analysis. Water Air Soil Pollut 201:195–208
Moberly JG, Staven A, Sani RK, Peyton BM (2010) Influence of pH and inorganic phosphate on toxicity of zinc to Arthrobacter sp. isolated from heavy-metal contaminated sediments. Environ Sci Technol. doi:10.1021/es100117f, In press
Mori K, Yamamoto H, Kamagata Y, Hatsu M, Takamizawa K (2000) Methanocalculus pumilus sp. nov., a heavy-metal-tolerant methanogen isolated from a waste-disposal site. Int J Syst Evol Microbiol 50:1723–1729
Nevin KP, Finneran KT, Lovley DR (2003) Microorganisms associated with uranium bioremediation in a high-salinity subsurface sediment. Appl Environ Microbiol 69:3672–3675
Niggemyer A, Spring S, Stackebrandt E, Rosenzweig RF (2001) Isolation and characterization of a novel As(V)-reducing bacterium: implications for arsenic mobilization and the genus Desulfitobacterium. Appl Environ Microbiol 67:5568–5580
Nold SC, Zhou J, Devol AH, Tiedje JM (2000) Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the beta subdivision of the Proteobacteria. Appl Environ Microbiol 66:4532–4535
Petrie L, North NN, Dollhopf SL, Balkwill DL, Kostka JE (2003) Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl Environ Microbiol 69:7467–7479
Purdy KJ, Nedwell DB, Embley TM (2003) Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl Environ Microbiol 69:3181–3191
Ramamoorthy S, Piotrowski JS, Langner HW, Holben WE, Morra MJ, Rosenzweig RF (2009) Ecology of sulfate-reducing bacteria in an iron-dominated, mining-impacted freshwater sediment. J Environ Qual 38:675–684
Rastogi G, Osman S, Kukkadapu R, Engelhard M, Vaishampayan PA, Andersen GL, Sani RK (2010) Microbial and mineralogical characterizations of soils collected from the deep biosphere of the former Homestake Gold Mine, South Dakota. Microb Ecol. (In press. doi:10.1007/s00248-010-9657-y)
Rastogi G, Osman S, Vaishampayan PA, Andersen GL, Stetler LD, Sani RK (2010) Microbial diversity in uranium mining-impacted soils as revealed by high-density 16S microarray and clone library. Microb Ecol 59:94–108
Rastogi G, Ranade DR, Yeole TY, Patole MS, Shouche YS (2008) Investigation of methanogen population structure in biogas reactor by molecular characterization of methyl-coenzyme M reductase A (mcrA) genes. Bioresour Technol 99:5317–5326
Rastogi G, Sani RK, Peyton BM, Moberly JG, Ginn TR (2009) Molecular studies on the microbial diversity associated with mining-impacted Coeur d'Alene river sediments. Microb Ecol 58:129–139
Roane TM (1999) Lead resistance in two bacterial isolates from heavy metal-contaminated soils. Microb Ecol 37:218–224
Sani RK, Rastogi G, Moberly JG, Dohnalkova A, Ginn TR, Spycher N, Shende N, Peyton BM (2010) The toxicity of lead to Desulfovibrio desulfuricans G20 in the presence of goethite and quartz. J Basic Microbiol 50:160–170
Sass H, Ramamoorthy S, Yarwood C, Langner H, Schumann P, Kroppenstedt RM, Spring S, Rosenzweig RF (2009) Desulfovibrio idahonensis sp. nov., sulfate-reducing bacteria isolated from a metal(loid)-contaminated freshwater sediment. Int J Syst Evol Microbiol 59:2208–2214
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schloss PD, Handelsman J (2006) Toward a census of bacteria in soil. PLoS Comput Biol 2:e92
Sevinc Sengor S, Sengor S, Spycher NF, Sani GTR, RK PB (2007) Biogeochemical reactive–diffusive transport of heavy metals in Lake Coeur d'Alene sediments. Appl Geochem 22:2569–2594
Simankova MV, Parshina SN, Tourova TP, Kolganova TV, Zehnder AJ, Nozhevnikova AN (2001) Methanosarcina lacustris sp. nov., a new psychrotolerant methanogenic archaeon from anoxic lake sediments. Syst Appl Microbiol 24:362–367
Sprenke KF, Rember WC, Bender SF, Hoffman ML, Rabbi F, Chamberlain VE (2000) Toxic metal contamination in the lateral lakes of the Coeur d'Alene River valley, Idaho. Environ Geol 39:575–586
Stein LY, La Duc MT, Grundl TJ, Nealson KH (2001) Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ Microbiol 3:10–18
von Wintzingerode F, Göbel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229
Wang H, Wang FY, Wei ZQ, Hu HY (2010) Quinone profiles of microbial communities in sediments of Haihe River-Bohai Bay as influenced by heavy metals and environmental factors. Environ Monit Assess. doi:10.1007/s10661-010-1573-6
Acknowledgments
The authors gratefully acknowledge the financial support provided by the National Science Foundation (grant #0628258) and Inland Northwest Research Alliance (INRA) Subsurface Science Graduate Fellowship program. Special thanks to Yvette M. Piceno and Gary L. Andersen at Lawrence Berkeley National Laboratory, CA for the microarray analysis. We also would like to thank the anonymous reviewers whose critiques were instrumental in improving the quality of our manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary class Table 1
Bacterial classes detected in CdAR sediments using PhyloChip analyses (DOC 67 kb)
Supplementary order Table 2
Bacterial orders detected in CdAR sediments using PhyloChip analyses (DOC 104 kb)
Supp. family Table 3
Bacterial families detected in CdAR sediments using PhyloChip analyses (DOC 150 kb)
Supp. genera Table 4
Bacterial genera retrieved on PhyloChip from CdAR sediments (DOC 1290 kb)
Rights and permissions
About this article
Cite this article
Rastogi, G., Barua, S., Sani, R.K. et al. Investigation of Microbial Populations in the Extremely Metal-Contaminated Coeur d'Alene River Sediments. Microb Ecol 62, 1–13 (2011). https://doi.org/10.1007/s00248-011-9810-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-011-9810-2