Abstract
Within the same human gastrointestinal tract, substantial differences in the bacterial species that inhabit oral cavity and intestinal tract have been noted. Previous research primarily attributed the differences to the influences of host environments and nutritional availabilities (“host habitat” effect). Our recent study indicated that, other than the host habitat effect, an existing microbial community could impose a selective pressure on incoming foreign bacterial species independent of host-mediated selection (“community selection” effect). In this study, we employed in vitro microbial floras representing microorganisms that inhabit the oral cavities and intestinal tract of mice in combination with Escherichia coli as a model intestinal bacterium and demonstrated that E. coli displays a striking community preference. It thrived when introduced into the intestinal microbial community and survived poorly in the microbial flora of foreign origin (oral community). A more detailed examination of this phenomenon showed that the oral community produced oxygen-free radicals in the presence of wild-type E. coli while mutants deficient in lipopolysaccharides (LPS) did not trigger significant production of these cell-damaging agents. Furthermore, mutants of E. coli defective in the oxidative stress response experienced a more drastic reduction in viability when cocultivated with the oral flora, while the exogenous addition of the antioxidant vitamin C was able to rescue it. We concluded that the oral-derived microbial community senses the E. coli LPS and kills the bacterium with oxygen-free radicals. This study reveals a new mechanism of community invasion resistance employed by established microflora to defend their domains.
Similar content being viewed by others
Introduction
The human gastrointestinal (GI) tract harbors trillions of bacterial cells and is one of the most complex ecosystems ever identified [30]. So far, over 700 and 1,000 bacterial species, respectively, have been identified in the oral cavity and intestinal tract, two of the distinct microbial niches along the GI tract [27, 30]. These dynamic resident microbiotas have important metabolic, trophic, and protective functions and greatly affect the host’s physiology and pathology [11]. Interestingly, despite their repeated exposure to a multitude of similar species, substantial differences in the bacterial composition between these two niches have been noted [24].
One of the intriguing questions in gastrointestinal tract-associated microbial community research is how the microbiota is formed and maintained [20]. Several factors have been proposed to play roles in establishing the microbial community structures in the niches provided by the human body. One of these factors is the “legacy effect”, which refers to the microbial composition in the local environment, or the inheritance of microflora from a parent [20]. Recent culture-independent microbial community analyses of human and mice gut revealed that the majority (>90%) of the bacterial phylotypes detected belong to the phyla Firmicutes and Bacteroidetes [19], while surveys of different fish species identified Proteobacteria as the most dominant bacterial phylum within their respective gut communities [13, 33]. The drastic microbial profile difference between mammals and fish could be due to the distinct microbial compositions they encounter in their different living environments. Another important factor in determining the GI tract-associated microbial community is the “host habitat” effect, or the distinct selective pressure asserted by the specific microenvironment within the host [32].
In a recent study, we cultivated bacterial mixtures from the GI tract of mice to establish stable in vitro oral and intestinal microbial communities, which contained at least 20 and 18 distinct bacterial species, respectively. Using this in vitro system, we demonstrated that bacterial species isolated from two different locations within the same GI tract (oral cavity or intestinal tract) are only compatible with bacterial communities that match their origins but are restricted by the respective “foreign” communities. The study suggested that, other than the “legacy” and “host habitat” effects, an existing microbial community could impose a selective pressure on incoming foreign bacterial species independent of host selection and might play an important role in restricting the integration of foreign bacteria and maintaining the stability of the existing community (“community selection” effect). The fact that most of the tested isolates failed to establish themselves in a foreign community also suggested an intriguing similarity to the invasion resistance effect experienced by a foreign species when it was trying to invade an established community in a nonmicrobial ecosystem [5, 7, 9]. In this study, we aim to further understand the underlying molecular mechanisms of the species restriction phenomenon between microbial communities of different origin.
Materials and Methods
Bacterial Strains and Growth Conditions
Escherichia coli strains, oral isolates, and bacterial mixtures were grown in brain heart infusion (BHI) broth supplemented with hemin (5 μg/ml), vitamin K (0.5 μg/ml), sucrose (0.1%), mannose (0.1%), and glucose (0.1%; simply referred to as BHI in this study), and cultures were incubated at 37 °C under microaerobic conditions (nitrogen 90%, carbon dioxide 5%, oxygen 5%) until turbid. When needed, kanamycin (30 μg/ml) was added to the medium for selecting E. coli.
Community Integration Assay
The preparation and establishment of the BHI cultivable intestinal microbial community (I-mix) and oral microbial community (O-mix) from the GI tract of mice were performed as previously described (see the accompanying manuscript). Briefly, overnight cultures of oral and intestinal microbial mixtures were seeded into six-well culture plates or 15 ml conical tubes containing 1.5 ml supplemented BHI medium. Cultures were incubated at 37 °C under microaerobic condition overnight to allow the establishment of the oral community as biofilm and intestinal community in pellet form to mimic high bacterial densities observed in oral and intestinal communities [10, 22].
PCR-denaturing gradient gel electrophoresis (PCR-DGGE) and viability count analysis were used to monitor the community integration assay (as described in the accompanying manuscript). Briefly, for PCR-DGGE analysis, an overnight culture of the original intestinal isolate (E. coli II-102) was adjusted to an OD600 nm of about 1 and added to the O-mix or I-mix, with a 1:10 ratio in cell numbers. Cocultivation samples were taken every 2 days, bacterial cells were treated with ethidium monoazide bromide (as described in the accompanying manuscript), collected by centrifugation, and total genomic DNA was isolated for further PCR-DGGE analysis to monitor the status of the isolate within the bacterial community. Two biological replicates were performed for each PCR-DGGE assay.
For viability count analysis, an E. coli strain (E. coli MG1655) carrying a kanamycin resistance marker was used as representative intestinal bacterial species in the community integration assay as described above. Cocultivation samples were taken periodically, subjected to serial dilutions and plated onto selective and nonselective supplemented BHI agar plates. Plates were incubated for 3 days at 37 °C under microaerobic condition before colonies were counted. Three replicates were performed for each viability count assay.
PCR-DGGE
PCR-DGGE was performed as described previously by Li et al. [21]. Briefly, the universal primer set, Bac1 and Bac2, was used to amply an approximately 300-bp internal fragment of the 16s rRNA gene. Each 50-μl PCR reaction contains 100 ng of purified genomic DNA, 40 pmol of each primer, 200 μM of each dNTP, 4.0 mM MgCl2, 5 μl of 10× PCR buffer, and 2.5 U of Taq DNA polymerase (Invitrogen). Cycling conditions were 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 56 °C for 1 min, and 72 °C for 2 min, with a final extension period of 5 min at 72 °C. The resulting PCR products were evaluated by electrophoresis in 1.0% agarose gels.
DGGE was performed using the D-Code gel system (Bio-Rad laboratories, Inc., Hercules, CA, USA). Approximately 300 ng of PCR products of each sample was resolved on 8% (w/v) polyacrylamide gels in 1× TAE buffer using denaturing gradients ranging from 40% to 70% (where 100% denaturant contains 7 M urea and 40% formamide). Electrophoresis was carried out at a fixed voltage (60 V) for 17 h. Electrophoresis buffer was maintained at 58 °C. Gels were stained with 1× TAE buffer containing 0.5 μg/ml ethidium bromide, and DGGE profile images were digitally recorded using the Molecular Imager Gel Documentation system (Bio-Rad Laboratories, Hercules, CA, USA). Diversity Database Software (Bio-Rad) was used to assess the change in the relative intensity of bands corresponding to bacterial species of interest.
Hydrogen Peroxide Production Assay
O-mix, original oral isolate Streptococcus salivarius-OI101, E. coli wild type (E. coli MG1655), and mutant strains were grown to the logarithmic phase, and cells were harvested and resuspended in fresh BHI medium to a final OD600 of 1. The O-mix was mixed with different bacterial strains at a 10:1 ratio or was mixed with purified lipopolysaccharide (LPS) from wild-type E. coli.
The hydrogen peroxide production assay was performed as previously described [17]. Briefly, spots of 5 μl of horseradish peroxidase (1 mg/ml; Thermo Scientific, Rockford, IL, USA) were added onto BHI agar plate containing 1 mg/ml leuco crystal violet (Acros Organics, Fair Lawn, NJ, USA). After the liquid was absorbed into the agar, 5 μl of the different mixtures was inoculated on top of the spots containing peroxidase. Plates were incubated at 37 °C under microaerobic condition. After overnight incubation, the plate was inspected for the development of a purple color on and around the colony. Three replicates were performed for this assay.
Hydrogen Peroxide Susceptibility Tests
The minimal inhibitory concentrations (MICs) of hydrogen peroxide were determined for each isolate by a dilution method using BHI medium supplemented with hydrogen peroxide at concentrations between 0 and 5 mM. Exponentially growing cultures of each isolate were harvested, resuspended, diluted to 106 cfu/ml in fresh BHI medium, and seeded into 96-well plates containing different concentrations of hydrogen peroxide. Plates were incubated at 37 °C overnight. MIC was defined as the lowest concentration that inhibited visible growth of bacteria. Three replicates were performed for each test.
E. coli Cell Membrane Preparation
Exponentially growing E. coli cells were harvested, and the cell pellet was washed two times with PBS and resuspended in lysis buffer (50 mM Tris–Cl, pH 8.0, 10 mM β-mercaptoethanol, 1 mM EDTA, and 50 mM NaCl). Cells were passed three times through a French Press at 20,000 psi. The cell lysate was centrifuged at 47,800×g for 30 min at 4 °C to pellet the membrane. The pellet was resuspended in lysis buffer and stored at −80 °C.
Heat, Fixation, and Pronase Treatments of E. coli Cells
Exponentially growing E. coli cells were harvested, washed two times, resuspended in PBS to an OD600 nm of 1 prior to the following different treatments: (1) heat treatment: Cells were incubated at 90 °C for 15 min and (2) fixation and pronase treatment: Formaldehyde was added to the cell suspension to a final concentration of 2%. After a 10-min incubation at room temperature, cells were spun down, washed three times, and resuspended in PBS. Pronase was then added to the cell suspension to a final concentration of 2 mg/ml, and the sample was incubated at 37 °C for 1 h. After the above treatments, cells were spun down, washed three times, and resuspended in fresh PBS.
LPS Purification
E. coli strains were grown to exponential phase, cultures were harvested, and the LPS was isolated using the LPS Extraction kit (iNtRON Biotechnology, Inc., Boca Raton, FL, USA) following manufacturer’s instructions. LPS was resuspended in ddH2O, and the concentration was determined with the HEK-Blue™ LPS Detection Kit (InvivoGen, San Diego, CA, USA).
Two-Chamber Assay
O-mix and E. coli strains were grown to the exponential phase, harvested, and resuspended in fresh BHI medium. The O-mix was inoculated into the bottom chamber of a 12-well plate containing a 0.4-μm PET membrane insert (Millipore, Billerica, MA, USA), while wild-type E. coli was added to the top chamber (in a 5:1 O-mix-to-E. coli ratio). Alternatively, the O-mix, together with either live, heat-killed, fixed, and pronase-treated wild-type E. coli (in a 10:1 O-mix-to-treated E. coli ratio), or cell membrane fractions of E. coli were added to the bottom chamber, while wild-type E. coli was added to the top chamber. Induction of killing under each condition was monitored by performing viability counts of the wild-type E. coli in the upper chamber. Three replicates were performed for each two-chamber assay.
Screens for E. coli Mutants with Reduced Ability in Inducing Killing and Altered Sensitivity to the Killing
Using the two-chamber assay setup, the E. coli Transposon Insertion Mutation and Large-scale Chromosome Deletion Libraries (NBRP, National Institute of Genetics, Japan) were screened for mutant derivatives with altered killing induction or survival in the presence of the O-mix.
Both O-mix and E. coli strains were grown to the exponential phase, harvested, and resuspended in fresh BHI medium. To screen for E. coli mutants with reduced killing-inducing ability, the O-mix, together with individual mutant strains (in a 10:1 O-mix-to-mutant ratio), was added to the bottom chamber of a 12-well plate containing a 0.4-μm PET membrane, while wild-type E. coli was added to the top chamber (in a 5:1 O-mix-to-wild-type ratio). When screening for mutants with altered sensitivity to the induced killing, the individual E. coli mutant strains were added to the top chamber, while E. coli wild type, together with O-mix, was added to the lower chamber. Viability of E. coli on the top chamber was monitored every 48 h by viability count.
Statistical Analysis
Significance of differences between average values was analyzed by t tests using MS Excel.
Results
The Intestinal Bacterium E. coli was Able to Integrate into Intestinal Community, While Being Excluded from Oral Community
In a recent study, we established stable microbial communities representing microorganisms that inhabit the oral cavity and intestinal tract using bacteria subcultured from mouse gastrointestinal tract. We found that oral and intestinal bacterial species display striking community preference, and they thrived when introduced into their bacterial community of origin and faired poorly in microbial flora of foreign origin. These results suggested that oral- and intestinal-derived bacteria community have a mechanism to restrict “foreign” intruders independent of host factor. To further investigate the molecular basis behind this species restriction phenomenon, we chose to study the fate of E. coli (an intestinal bacterium) when introduced into an oral microflora environment.
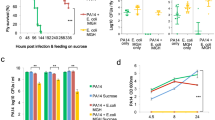
To test the compatibility of E. coli with the oral and intestinal communities, we performed a community preference assay. An intestinal isolate of E. coli (strain E. coli-II102) was added to the preformed oral (O-mix) and intestinal microbial communities (I-mix), respectively, in a ratio of 1: 10, and mixed cultures were incubated under microaerobic conditions. Samples were taken every 48 h and subjected to PCR-DGGE analysis. Intriguingly, in the presence of the O-mix, the intensity of the E. coli-associated band declined over time to barely detectable levels after 6 days while it remained prominent in the I-mix cultures (Fig. 1).
PCR-DGGE analysis of the community preference assay. Intestinal isolate, E. coli-II102, was cocultivated with either oral (O-mix) or intestinal (I-mix) communities in a 1:10 ratio. E. coli and whole community profiles were monitored by PCR-DGGE over a 6-day period. Two biological replicates were performed for each assay, and a representative gel image is shown
For a more quantitative analysis, E. coli strain MG1655 which carries a kanamycin resistance marker was chosen for the community preference assay. The viability count was monitored for both E. coli and the whole communities. Consistent with our DGGE analysis, E. coli MG1655 suffered a drastic loss in viability (105-fold) when cocultured with O-mix cells (P < 0.05; Fig. 2a), but little or no loss when cocultured with I-mix cells (Fig. 2b). The results confirmed that E. coli is compatible with intestinal bacterial communities that match its origins but is restricted by the oral communities. Since both the O-mix and I-mix communities were maintained in culture under the same nutritional conditions, the observed killing effects were likely originated from the preexisting microbial communities themselves rather than the environment.
Survival of E. coli in O-mix cocultures. a Wild-type E. coli was added to the established oral microbial community (O-mix) in a 1:10 ratio. Viability was monitored every 48 h for E. coli (open bars) and O-mix (solid bars); b wild-type E. coli was added to the established intestinal microbial community (I-mix) in a 1:10 ratio. Viability was monitored for E. coli (open bars) and I-mix (solid gray bars); c two-chamber assay in which E. coli was inoculated into the upper chamber and physically separated from O-mix in the lower chamber by a 0.4-μm pore-size membrane. Viability of E. coli (open bars) and O-mix (solid bars) was tracked by determining viable counts. d Two-chamber assay in which E. coli was added to the upper chamber, while O-mix, together with E. coli (in a 10:1 ratio) was inoculated into the lower chamber. Viability was monitored for O-mix and E. coli in both chambers (black solid bars represent total viable count of O-mix, gray solid bars and open bars represent viable count of E. coli in the lower and upper chamber, respectively). Three replicates were performed for each assay. Average values ± SD are shown. Star indicates that value of that time point is significantly lower than that of day 0 (P < 0.05)
O-mix Cells Generated Diffusible Killing Molecule(s) upon Physical Contact with E. coli
To determine if the observed killing requires direct cell–cell contact, we separated the E. coli and O-mix cells in a two-chamber vessel separated by a 0.4-μm membrane. As illustrated in Fig. 2c (inset), O-mix cells were incubated in the lower chamber, and E. coli cells were incubated in the upper chamber. Interestingly, E. coli was not killed by the O-mix cells when the cells shared the same culture medium but were physically separated by a membrane (Fig. 2c). However, when the O-mix cells and E. coli cells were allowed to make contact in the lower chamber, the E. coli cells in both chambers were killed (Fig. 2d). The result indicates that the O-mix cells generate (a) diffusible killing molecule(s) upon physical contact with E. coli cells or a component on the surface of E. coli cells, and the triggering component(s) are likely to be either surface proteins or exopolysaccharides. In an effort to differentiate between these possibilities, heat-killed, fixed, or pronase-treated E. coli or E. coli membrane fractions were added to the lower chamber of the two-chamber assay together with O-mix cells, and the viability of wild-type E. coli in the upper chamber was monitored periodically. All these different treatments failed to reduce the production of the killing factor (Supplemental Figure 1), indicating that the triggering components could be nonprotein components on the E. coli cell surface, such as lipopolysaccharides.
E coli Mutants Deficient in Exopolysaccharide Production Displayed Reduced Triggering Ability
To investigate which E. coli functions might trigger the killing response by the O-mix, we used the two-chamber assay to screen for mutants that display reduced triggering ability. For these experiments, we used previously described E. coli chromosomal deletion and transposon insertion mutant libraries [2, 23]. We first screened the large chromosomal deletion library to narrow down the region which might contain the genes of interest and perform further detailed screening for transposon insertion mutants within that region. Specifically, we added O-mix cells and mutant E. coli cells to the lower chamber and monitored viability of wild-type E. coli in the upper chamber. Our library screen led to the discovery of several mutant strains with reduced stimulation of the O-mix killing factor: the respective mutations mapped in genes that are either involved in LPS and enterobacterial common antigen (ECA) biosynthesis (e.g., waaL), or in modulating the polysaccharide chain length of LPS or ECA, such as wzzB and wzzE (Fig. 3a). Based on the putative functions encoded by these genes and the observation that the E. coli cell membrane fraction retained the killing factor triggering ability (Supplemental Figure 1), it was likely that the LPS or other exopolysaccharides of E. coli were responsible for inducing the production of killing molecules by the O-mix. To confirm this, extracted LPS from wild-type E. coli was added to the lower chamber which contained O-mix and the waaL mutant, and the viability of wild-type E. coli in the upper chamber was monitored periodically. The waaL mutant was less capable of inducing O-mix killing than the wild type; however, this triggering ability was partially restored when wild-type E. coli LPS was added to the experimental setup (Fig. 3b).
Analysis of E. coli mutants that show reduced stimulation of O-mix cell killing. In the two-chamber assay, a wild-type E. coli was added to the top chamber, while wt E. coli (black bar), E. coli-wzzB (gray bar), or E. coli-waaL (open bar) was added, together with the O-mix, to the bottom chamber. The viability of wild-type E. coli in the upper chamber of each well was monitored by viability count. b Wild-type E coli was added to the upper chamber, while O-mix, together with wild-type E. coli (black bars), or E. coli-waaL (gray bars), or E. coli-waaL plus LPS isolated from wild-type E. coli (open bars), were added to the lower chamber; viability of the wild-type E. coli in the upper chamber was monitored for each setup at indicated time intervals. Three replicates were performed for each assay. Average values ± SD are shown. Star indicates significant difference between the two values (P < 0.05)
E. coli Mutants Defective in Oxidative Stress Response Were More Sensitive to Killing
In order to further investigate which E. coli functions might render E. coli more sensitive or resistant to the killing factor, we performed the two-chamber assay to screen for mutants that display altered sensitivity to the killing. We added O-mix cells and wild-type cells to the lower chamber and screened the mutants in the upper chamber by monitoring their viability. A few mutants were identified in the screen and they mapped to two loci: One mutant was defective in the cadA/cadB, genes that are involved in production of cadaverine, the polyamine that has been suggested to protect E. coli cells from the toxic effect of oxygen; deletion in these genes caused an increased sensitivity to the killing factor (Fig. 4a). A second mutation mapped to katG, a locus that affects synthesis of the bifunctional catalase–peroxidase hydroperoxidase I in E. coli. Mutations in katG gene also rendered E. coli more susceptible to the killing factor (P < 0.05; Fig. 4b). Since both the cadA/cadB and katG loci have been shown to be involved in cellular defense against oxidative stress [6, 15, 16, 37], we speculated that the killing effect exerted by the oral community is likely related to oxidative damage.
Analysis of E. coli mutants that show increased sensitivity to the O-mix-derived killing factor. Using the two-chamber assay, a wild-type E. coli, together with O-mix, was added to the bottom chamber, while wild-type E. coli (black bar), E. coli-cadA (gray bar), or E. coli-cadB (open bar) was added to the top chamber. Viability of the E. coli in the upper chamber was monitored. b Wild-type E. coli, together with O-mix, was added to the bottom chamber, while wild-type E. coli (black bar), E. coli-katG (gray bar), or E. coli-oxyR (open bar) was added to the top chamber. Viability of the E. coli in the upper chamber was monitored. Three replicates were performed for each assay. Average values ± SD are shown. Star indicates significant difference between the two values (P < 0.05)
To further test this hypothesis, we chose E. coli-oxyR, a mutant defective in oxidative stress response due to a transposon insertional mutation in oxyR, for additional analysis. The oxyR gene encodes a transcriptional regulator that mediates the cellular oxidative stress defense. Consistent with our hypothesis, the two-chamber assay revealed that the oxyR mutant displayed increased sensitivity to the killing factor produced by the O-mix (Fig. 4b).
E. coli LPS Triggered Hydrogen Peroxide Production by the O-mix Cells
Since above mutant screens suggested that oxidative damage is involved in the killing of E. coli by O-mix cells, we examined oxygen-free radical production by the O-mix cells. As shown in Fig. 5, spot b2, O-mix cells produced very low amounts of oxygen-free radicals when grown on BHI agar plates containing leuco crystal violet and horseradish peroxidase. In the presence of the oral species S. salivarius, no increase in oxygen-free radicals by the O-mix cells was observed (Fig. 5, spot b3). However, co-incubation of the O-mix cells with wild-type E. coli cells triggered a dramatic increase in oxygen-free radical production as indicated by the purple color (Fig. 5, spot b1). This increase was not apparent when the O-mix cells were cocultured with E. coli mutants defective in LPS production (Fig 5, spots c1 and c2). Conversely, purified LPS (from wild-type E. coli) was sufficient to induce production of oxygen-free radicals (spot c3).
E. coli LPS triggers the release of oxygen-free radical by O-mix cells. a Wild-type E. coli alone (a1), O-mix, either alone (b2), mixed with the oral isolate (S. salivarius-OI100; b3), wild-type E. coli (b1), E. coli-waaL (c1), E. coli-wzzE (c2), or mixed with LPS isolated from wild-type E. coli (c3) was spotted onto BHI agar plate containing leuco crystal violet and horseradish peroxidase; purple color development was monitored after overnight incubation. Three replicates were performed and a representative result is shown
E. coli Was More Susceptible to Hydrogen Peroxide than Many Oral Isolates
In order to confirm that the intestinal isolate E. coli is more susceptible to hydrogen peroxide compared with oral bacteria, we performed an MIC assay. Results showed a MIC of 0.8 mM against E. coli, while most oral isolates displayed higher resistance to H2O2 up to 3 mM. Furthermore, oral mixture alone produced very low level of hydrogen peroxide, while the presence of E. coli significantly induced its hydrogen peroxide production (Supplementary Table 1).
Vitamin C (l-Ascorbic Acid) Offered Protection to E. coli Cells Against the Killing by O-mix
Above data suggested that hydrogen peroxide is likely the main factor responsible for the killing of the intestinal species E. coli by an oral bacterial community. To confirm this finding, vitamin C, an effective antioxidant, was added to the medium to test its ability to rescue E. coli from the oxidative stress-induced killing. In agreement with other results of this study, E. coli retained a significantly higher viability count (P < 0.05) in the presence of vitamin C, indicating that vitamin C conferred protection of E. coli cells against killing during cocultivation with the oral community (Fig. 6).
Vitamin C protection assay. Using the two-chamber assay, wild-type E. coli was added to the upper chamber, while O-mix, together with wild-type E. coli (dark bars) or wild-type E. coli plus 1 mM vitamin C (open bars), was added to the lower chamber. Viability of E. coli in the upper chamber (represented by different bars) was monitored for each setup. Three replicates were performed. Average values ± SD are shown. Star indicates significant difference between the two values (P < 0.05)
Discussion
In a recent study, we used in vitro microbial communities isolated from different parts of the GI tract of mice (oral cavity and intestines) as a model system to examine the interflora interactions. We demonstrated that an in vitro community composed of bacterial members isolated from the same natural niche exhibited invasion resistance and prevented the integration of bacterium of foreign origin (accompanying manuscript). This study was designed to further understand the underlying mechanisms of the species restriction phenomenon between microbial communities of different origin through molecular analysis using E. coli as a model intestinal bacterium.
E. coli was one of the bacterial species frequently isolated from the intestinal tract of mice, but not the oral cavity. As an intestinal bacterium, E. coli suffered great loss in viability when encountering an established oral microbial community (Fig. 1). This is in agreement with the result of our accompanying study in which all tested microbial isolates from the GI tract of mice displayed a striking community preference, and they only thrived in the community of their origin and were excluded from the community of foreign origin, suggesting that an existing microbial community could develop invasion resistance and impose selective pressure on incoming foreign bacterial species independent of host selection (“community selection” effect). From an ecological point of view, the invasion resistance is an important feature of established communities and is regarded as one of the key mechanisms in maintaining community stability [5]. The invasion resistance has been demonstrated in a variety of ecosystems, such as grassland [12], a marine ecosystem [36], and a stream fish community [3]. The fact that E. coli was unable to establish itself in the oral community suggested a similar invasion resistance effect against E. coli.
Although most of the current invasion resistance concepts were derived from nonmicrobial ecosystem, the striking similarity between the resistance phenomenon observed in the oral microbial community and nonmicrobial systems suggested that the similar principle could apply to the microbial world. First, we observed that the whole cultivable microbial communities are more effective than a community composed of fewer microbial species in excluding the integration of bacterium of different origin (accompanying manuscript). This is consistent with the observation that species-rich communities are more resistant to invasion by exotics than their species-poor counterparts [8]. Furthermore, our observation is corroborated by similar phenomena that were reported in other microbial community. Burmølle et al. demonstrated that a microbial community inhabiting the surface of the marine algae Ulva australis can inhibit the settlement of fouling organisms, and species isolated from the same epiphytic bacterial community can interact synergistically in biofilms and resist foreign bacterial invasion to a greater extent than single-species biofilms [4, 31]. More relevantly, it has been shown that the GI tract-associated indigenous microbiota can prevent the colonization of pathogens [35]. However, although these phenomena are well documented and several studies indicated that commensal bacterial isolates can produce antimicrobial compounds which could inhibit the growth of certain pathogens, no detailed analysis has been performed to further study the molecular mechanism(s) underlying this invasion resistance phenomenon between established microbial communities and invading species.
Using our established in vitro system, we demonstrated that the oral microbial community produced hydrogen peroxide in response to the presence of an intestinal bacterial species (E. coli) as an effective weapon to kill this foreign invader. Hydrogen peroxide is an effective bactericidal agent that is naturally produced by some organisms as a byproduct of oxygen metabolism. Commensal oral bacterial species, such as Streptococci spp. and Lactobacillus spp., are able to produce hydrogen peroxide [14, 17, 38, 39]; their protective role in defending their natural niches and protecting against infectious diseases has been suggested. For example, Uehara et al. demonstrated that viridans group streptococci may prevent methicillin-resistant Staphylococcus aureus colonization of the oral cavities of newborns via the production of hydrogen peroxide [38].
From the community point of view, the production of hydrogen peroxide could be a double-edged sword, while it kills the intruder, and it might also have negative effects on the residents of the community. However, our data suggested that the oral flora has a few mechanisms to keep this weapon in check. First, hydrogen peroxide is only produced when the oral community encounters intestinal invaders—E. coli cells (Fig. 2)—which ensures that the “weapon” is activated only when needed. Second, while the viability of E. coli suffered a drastic reduction during cocultivation, the viable count of oral microbial community remained quite constant (Fig. 2), and the profile of oral flora showed no significant change even after 4 days of cocultivation (Fig. 1), indicating that, compared to oral bacteria, E. coli might have a lower H2O2 tolerance. This is supported by the MIC data showing a MIC for hydrogen peroxide of less than 1 mM for E. coli, while most oral isolates displayed higher resistance to H2O2 up to 3 mM (Supplementary Table 1).
The fact that exogenously added vitamin C was able to rescue E. coli from being killed when cocultivated with the oral community further confirmed that hydrogen peroxide could be the main factor involved in killing intestinal species. Vitamin C—or ascorbic acid—is a reducing agent and well known for its antioxidant activity [26]. By providing electrons to reactive oxygen species, such as hydroxyl radicals formed from hydrogen peroxide, ascorbate is oxidized to dehydroascorbate, which is relatively stable and does not cause cell damage, while the reactive oxygen species can be reduced to water.
The most intriguing finding of our study is that the oral microbial community is able to detect the presence of the intestinal bacterial species E. coli and trigger the hydrogen peroxide production defensive response by recognizing the E. coli LPS. LPS is a major component of the outer membrane of gram-negative bacteria, contributing to their structural integrity [29]. However, due to its exposure to the very outer surface of the bacterial cell, LPS is also an easy target for recognition by host antibodies and can be detected by an ancient receptor—toll-like receptor 4—of the innate immune system present on immune cells, such as macrophages and neutrophils [28, 34] and induce a strong response from normal animal immune systems [1, 25]. Upon recognizing bacterial LPS, one of the effectors produced by immune cells in an effort to eliminate and prevent the invasion of foreign bacteria is hydrogen peroxide [18, 34], the same toxic chemical we found to be produced by oral bacterial cells when physically interacting with E. coli.
The recognition of E. coli LPS by O-mix cells was correlated with the ability of these cells to kill E. coli since the E. coli LPS mutant (waaL) exhibited greatly reduced induction of oxygen-free radicals (Fig. 5, spot c1; and thus killing) than wild type; however, this triggering ability was restored when wild-type E. coli LPS was added to the medium (Fig. 3). Taken together, these results show that bacteria in the O-mix have the ability to recognize the LPS of E. coli and then trigger the production of oxygen-free radicals to kill E. coli, a mechanism similar to that employed by the body’s immune response. Further work is needed to identity the sensor component for the E. coli LPS and the corresponding hydrogen peroxide producer within the oral community.
In our in vitro system, a significant number of the intestinal microflora members belong to the Enterobacteriaceae family, including E. coli, Enterobacter spp., and Shigella spp., which are gram-negative species carrying LPS on their outer membrane, while the oral community is mainly comprised of gram-positive species, such as Streptococcus spp., Staphylococcus spp., and Lactobacillus spp. In the oral cavity, these gram-positive microbes are the major species detected in saliva and supragingival plaque where often the first encounter between the introduced intestinal bacteria and oral microbes takes place. It would be interesting to know whether the presence of LPS on the surface of other intestinal bacterial species can also trigger hydrogen peroxide production and if the LPS-induced H2O2 production is a more generalized mechanism employed by the oral community to prevent the integration of certain intestinal bacteria.
Due to the limitation of culture-dependent methods, the results we obtained using the in vitro system cannot entirely represent the real situation in native conditions. Information regarding the contributions of host selection and noncultivable subpopulation to the shaping and maintenance of the microbial community could not be obtained using in vitro system, and it is possible that factors other than induced oxidative stress could also be contributing to the killing of E. coli. Nevertheless, the results presented here provide important molecular insights as to the mechanism by which oral bacteria restrict colonization of the oral cavity by one of intestinal bacteria—E. coli. Understanding the mechanism of interfloral relationships is important as it will enhance our understanding of the data derived from the human microbiome project and assist in the development of new therapeutic strategies for modulating the protective/probiotic effects of different microbial communities against different pathogens.
References
Aderem A, Ulevitch RJ (2000) Toll-like receptors in the induction of the innate immune response. Nature 406:782–787
Baba T, Huan HC, Datsenko K, Wanner BL, Mori H (2008) The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12. Meth Mol Biol 416:183–194
Baltz DM, Moyle PB (1993) Invasion resistance to introduced species by a native assemblage of California stream fishes. Ecol Applications 3:246–255
Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S (2006) Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol 72:3916–3923
Case TJ (1990) Invasion resistance arises in strongly interacting species-rich model competition communities. Proc Natl Acad Sci USA 87:9610–9614
Chattopadhyay MK, Tabor CW, Tabor H (2003) Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc Natl Acad Sci USA 100:2261–2265
deRivera CE, Ruiz GM, Anson HH, Jivoff P (2005) Biotic resistance to invasion: native predator limits abundance and distribution of an introduced crab. Ecology 86:3364–3376
Elton CS (1958) The ecology of invasions by animals and plants. The University of Chicago Press, Chicago
Ford SD (1996) Range extension by the oyster parasite Perkinsus marinus into northeastern United States: response to climate change? J Shellfish Res 15:45–56
Gorbach S (1971) Intestinal microflora. Gastroenterology 60:1110–1129
Guarner F, Malagelada JR (2003) Gut flora in health and disease. The Lancet 361:512–519
Hector A, Dobson K, Minns A, Bazeley-White E, Lawton JH (2001) Community diversity and invasion resistance: an experimental test in a grassland ecosystem and a review of comparable studies. Ecol Res 16:819–831
Huber I, Spanggaard B, Appel KF, Rossen L, Nielsen T, Gram L (2004) Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout. J Appl Microbiol 96:117–132
Huichun T, Wei C, Justin M, Qi F, Shi W, Dong X (2007) Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mole Microbiol 63:872–880
Jung IL, Kim IJ (2003) Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem and Biophys Res Commun 301:915–922
Kim JS, Choi SH, Lee JK (2006) Lysine decarboxylase expression by Vibrio vulnificus is induced by SoxR in response to superoxide stress. J Bacteriol 188:8586–8592
Kreth J, Merritt J, Shi W, Qi F (2005) Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203
Landmann R, Scherer F, Schumann R, Link S, Sansano S, Zimmerli W (1995) LPS directly induces oxygen radical production in human monocytes via LPS binding protein and CD14. J Leukoc Biol 57:440–449
Ley RE, Beckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075
Ley RE, Peterson DA, Gordon JI (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848
Li Y, Ku CYS, Xu J, Saxena D, Caufield PW (2005) Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res 84:559–564
Lobene RR (1979) Clinical studies of plaque control agents: an overview. J Dent Res 58:2381–2388
Masayuki H, Toshiharu I, Hiroshi M, Kimie T, Kazuyuki F, Kenji K, Tomotake O, Takehiro Y, Yukiko Y, Hideo M, Tsutomu K, Jun-ichi K (2005) Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mole Microbiol 55:137–149
Maukonen J, Matto J, Suihko ML, Saarela M (2008) Intra-individual diversity and similarity of salivary and faecal microbiota. J Med Microbiol 57:1560–1568
Medzhitov R, Janeway C (2000) Innate immunity. N Engl J Med 343:338–344
Padayatty SJ, Katz A, Wang Y, Eck P, Kwon O, Lee JH, Chen S, Corpe C, Dutta A, Dutta SK, Levine M (2003) Vitamin C as an antioxidant: evaluation of its role in disease prevention. J Am Coll Nutr 22:18–35
Paster BJ, Olsen I, Aas JA, Dewhirst EF (2006) The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 42:80–87
Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085–2088
Raetz CRH, Whitfield C (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700
Rajilić-Stojanović M, Smidt H, de Vos W (2007) Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 9:2125–2136
Rao D, Webb JS, Holmström C, Case R, Low A, Steinberg P, Kjelleberg S (2007) Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl Environ Microbiol 73:7844–7852
Rawls JF, Mahowald MA, Ley RE, Gordon JI (2006) Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433
Rawls JF, Samuel BS, Gordon JI (2004) Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA 101:4596–4601
Remer KA, Brcic M, Jungi TW (2003) Toll-like receptor-4 is involved in eliciting an LPS-induced oxidative burst in neutrophils. Immunol Lett 85:75–80
Shimizu A, Shimizu K, Nakamura T (2008) Non-pathogenic bacterial flora may inhibit colonization by methicillin-resistant Staphylococcus aureus in extremely low birth weight infants. Neonatol 93:158–161
Stachowicz JJ, Fried H, Osman RW, Whitlatchd RB (2001) Biodiversity, invasion resistance, and marine ecosystem function: reconciling pattern and process. Ecology 83:2575–2590
Triggs-Raine BL, Doble BW, Mulvey MR, Sorby PA, Loewen PC (1988) Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J Bacteriol 170:4415–4419
Uehara Y, Kikuchi K, Nakamura T, Nakama H, Agematsu K, Kawakami Y, Maruchi N, Totsuka K (2001) H2O2 produced by viridans group streptococci may contribute to inhibition of methicillin resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin Infect Dis 32:1408–1413
Uehara Y, Kikuchi K, Nakamura T, Nakama H, Agematsu K, Kawakami Y, Maruchi N, Totsuka K (2001) Inhibition of methicillin-resistant Staphylococcus aureus colonization of oral cavities in newborns by viridans group streptococci. Clin Infect Dis 32:1399–1407
Acknowledgments
We thank T. Ano and Z. Yang for their help in the initial characterization of mice GI tract isolates; L. Li, W. Hu, and J. Zhou for discussions; and National BioResource Project (NIG, Japan) for providing E. coli mutant library. The study was supported in part by NIH GM54666 to WS and GM20509 to DRZ.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
(DOC 104 kb)
Supplementary Table 1
Susceptibility of bacterial isolates to hydrogen peroxide and hydrogen peroxide production by O-mix in the absence and presence of E. coli (DOC 101 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
He, X., Tian, Y., Guo, L. et al. Oral-Derived Bacterial Flora Defends Its Domain by Recognizing and Killing Intruders—A Molecular Analysis Using Escherichia coli as a Model Intestinal Bacterium. Microb Ecol 60, 655–664 (2010). https://doi.org/10.1007/s00248-010-9708-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9708-4