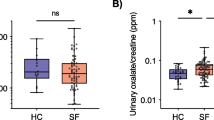
Abstract
Intestinal microbiome dysbiosis is a known risk factor for recurrent kidney stone disease (KSD) with prior data suggesting a role for dysfunctional metabolic pathways other than those directly utilizing oxalate. To identify alternative mechanisms, the current study analyzed differences in the metabolic potential of intestinal microbiomes of patients (n = 17) and live-in controls (n = 17) and determined their relevance to increased risk for KSD using shotgun metagenomic sequencing. We found no differences in the abundance of genes associated with known oxalate degradation pathways, supporting the notion that dysfunction in other metabolic pathways plays a role in KSD. Further analysis showed decreased abundance of key enzymes involved in butyrate biosynthesis in patient intestinal microbiomes. Furthermore, de novo construction of microbial genomes showed that the majority of genes significantly enriched in non-stone formers are affiliated with Faecalibacterium prausnitzii, a major butyrate producer. Specifically pertaining to butyrate metabolism, the majority of abundant genes mapped back to F. prausnitzii, Alistipes spp., and Akkermansia muciniphila. No differences were observed in ascorbate or glyoxylate metabolic pathways. Collectively, these data suggest that impaired bacterial-associated butyrate metabolism may be an oxalate-independent mechanism that contributes to an increased risk for recurrent KSD. This indicates that the role of the intestinal microbiome in recurrent KSD is multi-factorial, which is representative of the highly intertwined metabolic nature of this complex environment. Future bacteria-based treatments must not be restricted to targeting only oxalate metabolism.




Similar content being viewed by others
Data availability
Raw shotgun metagenomic data were deposited to the sequence read archive under accession # SRR15021121- SRR15021153.
References
Allison MJ, Dawson KA, Mayberry WR, Foss JG (1985) Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol 141(1):1–7. https://doi.org/10.1007/BF00446731
Hatch M, Cornelius J, Allison M, Sidhu H, Peck A, Freel RW (2006) Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int 69(4):691–698. https://doi.org/10.1038/sj.ki.5000162
Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW (2011) Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest Liver Physiol 300(3):G461–G469. https://doi.org/10.1152/ajpgi.00434.2010
Hatch M, Freel RW (2013) A human strain of Oxalobacter (HC-1) promotes enteric oxalate secretion in the small intestine of mice and reduces urinary oxalate excretion. Urolithiasis 41(5):379–384. https://doi.org/10.1007/s00240-013-0601-8
Arvans D, Jung YC, Antonopoulos D, Koval J, Granja I, Bashir M et al (2017) Oxalobacter formigenes-derived bioactive factors stimulate oxalate transport by intestinal epithelial cells. J Am Soc Nephrol 28(3):876–887. https://doi.org/10.1681/ASN.2016020132
Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM et al (2008) Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol 19(6):1197–1203. https://doi.org/10.1681/ASN.2007101058
Troxel SA, Sidhu H, Kaul P, Low RK (2003) Intestinal Oxalobacter formigenes colonization in calcium oxalate stone formers and its relation to urinary oxalate. J Endourol 17(3):173–176. https://doi.org/10.1089/089277903321618743
Tavasoli S, Alebouyeh M, Naji M, Shakiba Majd G, Shabani Nashtaei M, Broumandnia N et al (2020) Association of intestinal oxalate-degrading bacteria with recurrent calcium kidney stone formation and hyperoxaluria: a case-control study. BJU Int 125(1):133–143. https://doi.org/10.1111/bju.14840
Siener R, Bangen U, Sidhu H, Honow R, von Unruh G, Hesse A (2013) The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney Int 83(6):1144–1149. https://doi.org/10.1038/ki.2013.104
Sidhu H, Schmidt ME, Cornelius JG, Thamilselvan S, Khan SR, Hesse A et al (1999) Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract-dwelling bacterium Oxalobacter formigenes: possible prevention by gut recolonization or enzyme replacement therapy. J Am Soc Nephrol 10(Suppl 14):S334–S340
Kumar R, Ghoshal UC, Singh G, Mittal RD (2004) Infrequency of colonization with Oxalobacter formigenes in inflammatory bowel disease: possible role in renal stone formation. J Gastroenterol Hepatol 19(12):1403–1409. https://doi.org/10.1111/j.1440-1746.2004.03510.x
Sikora P, Niedzwiadek J, Mazur E, Paluch-Oles J, Zajaczkowska M, Koziol-Montewka M (2009) Intestinal colonization with Oxalobacter formigenes and its relation to urinary oxalate excretion in pediatric patients with idiopathic calcium urolithiasis. Arch Med Res 40(5):369–373. https://doi.org/10.1016/j.arcmed.2009.05.004
Liu M, Zhang Y, Wu J, Gao M, Zhu Z, Chen H (2023) Causal relationship between kidney stones and gut microbiota contributes to the gut-kidney axis: a two-sample Mendelian randomization study. Front Microbiol 14:1204311. https://doi.org/10.3389/fmicb.2023.1204311
Miller AW, Choy D, Penniston KL, Lange D (2019) Inhibition of urinary stone disease by a multi-species bacterial network ensures healthy oxalate homeostasis. Kidney Int 96(1):180–188. https://doi.org/10.1016/j.kint.2019.02.012
Denburg MR, Koepsell K, Lee JJ, Gerber J, Bittinger K, Tasian GE (2020) Perturbations of the gut microbiome and metabolome in children with calcium oxalate kidney stone disease. J Am Soc Nephrol 31(6):1358–1369. https://doi.org/10.1681/ASN.2019101131
Zampini A, Nguyen AH, Rose E, Monga M, Miller AW (2019) Defining dysbiosis in patients with urolithiasis. Sci Rep 9(1):5425. https://doi.org/10.1038/s41598-019-41977-6
Miller AW, Dearing D (2013) The metabolic and ecological interactions of oxalate-degrading bacteria in the Mammalian gut. Pathogens 2(4):636–652. https://doi.org/10.3390/pathogens2040636
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1):59–60
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):1
Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K et al (2016) MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102:3–11. https://doi.org/10.1016/j.ymeth.2016.02.020
Konwar KM, Hanson NW, Bhatia MP, Kim D, Wu SJ, Hahn AS et al (2015) MetaPathways v2.5: quantitative functional, taxonomic and usability improvements. Bioinformatics 31(20):3345–3347. https://doi.org/10.1093/bioinformatics/btv361
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30. https://doi.org/10.1093/nar/28.1.27
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628. https://doi.org/10.1038/nmeth.1226
Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN (2010) RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26(4):493–500. https://doi.org/10.1093/bioinformatics/btp692
Miller IJ, Rees ER, Ross J, Miller I, Baxa J, Lopera J et al (2019) Autometa: automated extraction of microbial genomes from individual shotgun metagenomes. Nucleic Acids Res 47(10):e57. https://doi.org/10.1093/nar/gkz148
Aganezov SS, Alekseyev MA (2017) CAMSA: a tool for comparative analysis and merging of scaffold assemblies. BMC Bioinform 18(15):41–50
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19(6):1117–1123
Wilcoxon F (1946) Individual comparisons of grouped data by ranking methods. J Econ Entomol 39:269. https://doi.org/10.1093/jee/39.2.269
R Core Team (2017) R: a language and environment for statistical computing. https://www.R-project.org/
Hothorn T, Hornik K, van de Wiel M, Zeileis A (2008) Implementing a class of permutation tests: the coin package. J Stat Softw. 28:1–23
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. (2022) Vegan: community ecology package. https://cran.r-project.org/web/packages/vegan/index.html
Anand S, Kaur H, Mande SS (2016) Comparative in silico analysis of butyrate production pathways in gut commensals and pathogens. Front Microbiol 7:1945. https://doi.org/10.3389/fmicb.2016.01945
Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK et al (2017) Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35(8):725–731. https://doi.org/10.1038/nbt.3893
Miller AW, Dale C, Dearing MD (2017) Microbiota diversification and crash induced by dietary oxalate in the mammalian herbivore Neotoma albigula. mSphere. https://doi.org/10.1128/mSphere.00428-17
Ferraro PM, Curhan GC, Gambaro G, Taylor EN (2019) Antibiotic use and risk of incident kidney stones in female nurses. Am J Kidney Dis 74(6):736–741. https://doi.org/10.1053/j.ajkd.2019.06.005
Tasian GE, Jemielita T, Goldfarb DS, Copelovitch L, Gerber JS, Wu Q et al (2018) Oral antibiotic exposure and kidney stone disease. J Am Soc Nephrol 29(6):1731–1740. https://doi.org/10.1681/ASN.2017111213
Wilkins LJ, Monga M, Miller AW (2019) Defining dysbiosis for a cluster of chronic diseases. Sci Rep 9(1):12918. https://doi.org/10.1038/s41598-019-49452-y
Shimizu S, Inoue K, Tani Y, Yamada H (1981) Butyryl-CoA synthetase of Pseudomonas aeruginosa–purification and characterization. Biochem Biophys Res Commun 103(4):1231–1237. https://doi.org/10.1016/0006-291x(81)90254-0
Tersteegen A, Linder D, Thauer RK, Hedderich R (1997) Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur J Biochem 244(3):862–868. https://doi.org/10.1111/j.1432-1033.1997.00862.x
Kletzin A, Adams MW (1996) Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol 178(1):248–257. https://doi.org/10.1128/jb.178.1.248-257.1996
Chabriere E, Charon MH, Volbeda A, Pieulle L, Hatchikian EC, Fontecilla-Camps JC (1999) Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol 6(2):182–190. https://doi.org/10.1038/5870
Furdui C, Ragsdale SW (2000) The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem 275(37):28494–28499. https://doi.org/10.1074/jbc.M003291200
Gibson MI, Brignole EJ, Pierce E, Can M, Ragsdale SW, Drennan CL (2015) The structure of an oxalate oxidoreductase provides insight into microbial 2-oxoacid metabolism. Biochemistry 54(26):4112–4120. https://doi.org/10.1021/acs.biochem.5b00521
Stern JM, Moazami S, Qiu Y, Kurland I, Chen Z, Agalliu I et al (2016) Evidence for a distinct gut microbiome in kidney stone formers compared to non-stone formers. Urolithiasis 44(5):399–407. https://doi.org/10.1007/s00240-016-0882-9
Chen F, Bao X, Liu S, Ye K, Xiang S, Yu L et al (2021) Gut microbiota affect the formation of calcium oxalate renal calculi caused by high daily tea consumption. Appl Microbiol Biotechnol 105(2):789–802. https://doi.org/10.1007/s00253-020-11086-w
Cui Q, Xia Y, Wu Q, Chang Q, Niu K, Zhao Y (2021) A meta-analysis of the reproducibility of food frequency questionnaires in nutritional epidemiological studies. Int J Behav Nutr Phys Act 18(1):12. https://doi.org/10.1186/s12966-020-01078-4
Archer E, Marlow ML, Lavie CJ (2018) Controversy and debate: memory-based methods paper 1: the fatal flaws of food frequency questionnaires and other memory-based dietary assessment methods. J Clin Epidemiol 104:113–124. https://doi.org/10.1016/j.jclinepi.2018.08.003
Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C et al (2000) Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 66(4):1654–1661. https://doi.org/10.1128/aem.66.4.1654-1661.2000
Duncan SH, Hold GL, Harmsen HJM, Stewart CS, Flint HJ (2002) Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 52(6):2141–2146. https://doi.org/10.1099/00207713-52-6-2141
Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M (2017) Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J 11(4):841–852. https://doi.org/10.1038/ismej.2016.176
Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ (2002) The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217(2):133–139. https://doi.org/10.1111/j.1574-6968.2002.tb11467.x
Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A (2013) Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog 5(1):23. https://doi.org/10.1186/1757-4749-5-23
Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J et al (2017) Microbial metabolic networks at the mucus layer lead to Diet-independent butyrate and vitamin B12 production by intestinal symbionts. MBio. https://doi.org/10.1128/mBio.00770-17
Ticinesi A, Milani C, Guerra A, Allegri F, Lauretani F, Nouvenne A et al (2018) Understanding the gut-kidney axis in nephrolithiasis: an analysis of the gut microbiota composition and functionality of stone formers. Gut 67(12):2097–2106. https://doi.org/10.1136/gutjnl-2017-315734
Al KF, Joris BR, Daisley BA, Chmiel JA, Bjazevic J, Reid G et al (2023) Multi-site microbiota alteration is a hallmark of kidney stone formation. Microbiome 11(1):263. https://doi.org/10.1186/s40168-023-01703-x
Moens F, De Vuyst L (2017) Inulin-type fructan degradation capacity of Clostridium cluster IV and XIVa butyrate-producing colon bacteria and their associated metabolic outcomes. Benef Microbes 8(3):473–490. https://doi.org/10.3920/BM2016.0142
Lee B, Moon KM, Kim CY (2018) Tight junction in the intestinal epithelium: its association with diseases and regulation by phytochemicals. J Immunol Res 2018:2645465. https://doi.org/10.1155/2018/2645465
Knauf F, Ko N, Jiang Z, Robertson WG, Van Itallie CM, Anderson JM et al (2011) Net intestinal transport of oxalate reflects passive absorption and SLC26A6-mediated secretion. J Am Soc Nephrol 22(12):2247–2255. https://doi.org/10.1681/ASN.2011040433
Whittamore JM, Hatch M (2017) The role of intestinal oxalate transport in hyperoxaluria and the formation of kidney stones in animals and man. Urolithiasis 45(1):89–108. https://doi.org/10.1007/s00240-016-0952-z
Farre R, Fiorani M, Abdu Rahiman S, Matteoli G (2020) Intestinal permeability, inflammation and the role of nutrients. Nutrients. https://doi.org/10.3390/nu12041185
Krishnan S, Rajendran VM, Binder HJ (2003) Apical NHE isoforms differentially regulate butyrate-stimulated Na absorption in rat distal colon. Am J Physiol Cell Physiol 285(5):C1246–C1254. https://doi.org/10.1152/ajpcell.00598.2002
Musch MW, Bookstein C, Xie Y, Sellin JH, Chang EB (2001) SCFA increase intestinal Na absorption by induction of NHE3 in rat colon and human intestinal C2/bbe cells. Am J Physiol Gastrointest Liver Physiol 280(4):G687–G693. https://doi.org/10.1152/ajpgi.2001.280.4.G687
Kiela PR, Hines ER, Collins JF, Ghishan FK (2001) Regulation of the rat NHE3 gene promoter by sodium butyrate. Am J Physiol Gastrointest Liver Physiol 281(4):G947–G956. https://doi.org/10.1152/ajpgi.2001.281.4.G947
Alrefai WA, Wen X, Jiang W, Katz JP, Steinbrecher KA, Cohen MB et al (2007) Molecular cloning and promoter analysis of downregulated in adenoma (DRA). Am J Physiol Gastrointest Liver Physiol 293(5):G923–G934. https://doi.org/10.1152/ajpgi.00029.2007
Canani RB, Terrin G, Elce A, Pezzella V, Heinz-Erian P, Pedrolli A et al (2013) Genotype-dependency of butyrate efficacy in children with congenital chloride diarrhea. Orphanet J Rare Dis 8:194. https://doi.org/10.1186/1750-1172-8-194
Freel RW, Whittamore JM, Hatch M (2013) Transcellular oxalate and Cl- absorption in mouse intestine is mediated by the DRA anion exchanger Slc26a3, and DRA deletion decreases urinary oxalate. Am J Physiol Gastrointest Liver Physiol 305(7):G520–G527. https://doi.org/10.1152/ajpgi.00167.2013
Freel RW, Hatch M, Green M, Soleimani M (2006) Ileal oxalate absorption and urinary oxalate excretion are enhanced in Slc26a6 null mice. Am J Physiol Gastrointest Liver Physiol 290(4):G719–G728. https://doi.org/10.1152/ajpgi.00481.2005
Jiang Z, Asplin JR, Evan AP, Rajendran VM, Velazquez H, Nottoli TP et al (2006) Calcium oxalate urolithiasis in mice lacking anion transporter Slc26a6. Nat Genet 38(4):474–478. https://doi.org/10.1038/ng1762
Andrade-Oliveira V, Amano MT, Correa-Costa M, Castoldi A, Felizardo RJ, de Almeida DC et al (2015) Gut Bacteria products prevent AKI induced by ischemia-reperfusion. J Am Soc Nephrol 26(8):1877–1888. https://doi.org/10.1681/ASN.2014030288
Machado RA, Constantino Lde S, Tomasi CD, Rojas HA, Vuolo FS, Vitto MF et al (2012) Sodium butyrate decreases the activation of NF-kappaB reducing inflammation and oxidative damage in the kidney of rats subjected to contrast-induced nephropathy. Nephrol Dial Transplant 27(8):3136–3140. https://doi.org/10.1093/ndt/gfr807
Al-Harbi NO, Nadeem A, Ahmad SF, Alotaibi MR, AlAsmari AF, Alanazi WA et al (2018) Short chain fatty acid, acetate ameliorates sepsis-induced acute kidney injury by inhibition of NADPH oxidase signaling in T cells. Int Immunopharmacol 58:24–31. https://doi.org/10.1016/j.intimp.2018.02.023
Marzocco S, Fazeli G, Di Micco L, Autore G, Adesso S, Dal Piaz F et al (2018) Supplementation of short-chain fatty acid, sodium propionate, in patients on maintenance hemodialysis: beneficial effects on inflammatory parameters and gut-derived uremic toxins, a pilot Study (PLAN Study). J Clin Med. https://doi.org/10.3390/jcm7100315
Yang J, Li Q, Henning SM, Zhong J, Hsu M, Lee R et al (2018) Effects of prebiotic fiber xylooligosaccharide in adenine-induced nephropathy in mice. Mol Nutr Food Res. https://doi.org/10.1002/mnfr.201800014
Vaziri ND, Liu SM, Lau WL, Khazaeli M, Nazertehrani S, Farzaneh SH et al (2014) High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS ONE 9(12):e114881. https://doi.org/10.1371/journal.pone.0114881
Wong J, Piceno YM, DeSantis TZ, Pahl M, Andersen GL, Vaziri ND (2014) Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am J Nephrol 39(3):230–237. https://doi.org/10.1159/000360010
Huang W, Zhou L, Guo H, Xu Y, Xu Y (2017) The role of short-chain fatty acids in kidney injury induced by gut-derived inflammatory response. Metabolism 68:20–30. https://doi.org/10.1016/j.metabol.2016.11.006
Khan SR (2013) Reactive oxygen species as the molecular modulators of calcium oxalate kidney stone formation: evidence from clinical and experimental investigations. J Urol 189(3):803–811. https://doi.org/10.1016/j.juro.2012.05.078
Khan SR, Canales BK, Dominguez-Gutierrez PR (2021) Randall’s plaque and calcium oxalate stone formation: role for immunity and inflammation. Nat Rev Nephrol 17(6):417–433. https://doi.org/10.1038/s41581-020-00392-1
Liu Y, Jin X, Hong HG, Xiang L, Jiang Q, Ma Y et al (2020) The relationship between gut microbiota and short chain fatty acids in the renal calcium oxalate stones disease. FASEB J 34(8):11200–11214. https://doi.org/10.1096/fj.202000786R
Liu Y, Jin X, Ma Y, Jian Z, Wei Z, Xiang L et al (2021) Short-chain fatty acids reduced renal calcium oxalate stones by regulating the expression of intestinal oxalate transporter SLC26A6. mSystems 6(6):e0104521. https://doi.org/10.1128/mSystems.01045-21
Hosomi A, Nakanishi T, Fujita T, Tamai I (2012) Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS ONE 7(2):e30456. https://doi.org/10.1371/journal.pone.0030456
Goncalves P, Gregorio I, Martel F (2011) The short-chain fatty acid butyrate is a substrate of breast cancer resistance protein. Am J Physiol Cell Physiol 301(5):C984–C994. https://doi.org/10.1152/ajpcell.00146.2011
Guo Z, Zhang J, Wang Z, Ang KY, Huang S, Hou Q et al (2016) Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep 6:20602. https://doi.org/10.1038/srep20602
Robijn S, Hoppe B, Vervaet BA, D’Haese PC, Verhulst A (2011) Hyperoxaluria: a gut-kidney axis? Kidney Int 80(11):1146–1158. https://doi.org/10.1038/ki.2011.287
Pearle MS (2003) Effect of vitamin C supplements on urinary oxalate and pH in calcium stone-forming patients. Int Braz J Urol 29(2):167–168
Funding
Canadian Urologic Association—Astellas Research Grant Awarded to Dirk Lange. The funding body had no role in the design of the study and collection, analysis and interpretation of data or the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
Conception/design of the work: BC, DL. Acquisition, analysis, interpretation of data: BC, DL, WHC, AA, CML, EKG, SH, ARM, AM, KP. Software used in the work: WHC, SH, CML, AKG, ARM, AM, AA. Drafting/revision of manuscript: all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests associated with this work.
Ethical approval and consent to participate
The study was approved by the University of British Columbia Clinical Research Ethics Board with informed consent obtained from all participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choy, W.H., Adler, A., Morgan-Lang, C. et al. Deficient butyrate metabolism in the intestinal microbiome is a potential risk factor for recurrent kidney stone disease. Urolithiasis 52, 38 (2024). https://doi.org/10.1007/s00240-024-01534-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00240-024-01534-x