Abstract
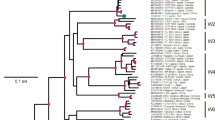
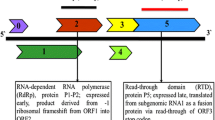
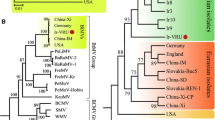
The complete genome of the German isolate AC of Apple stem grooving virus (ASGV) was sequenced. It encodes two overlapping open reading frames (ORFs), similarly to previously described ASGV isolates. Two regions of high variability were detected between the ASGV isolates, variable region 1 (V1, from amino acids (aa) 532 to 570), and variable region 2 (V2, from aa 1,583 to 1,868). The phylogenetic analysis of the V1 and V2 regions suggested that the ASGV diversity was structured by host plant species rather than geographical origin. The dN/dS ratio between nonsynonymous and synonymous nucleotide substitution rates varied greatly along the ASGV genome. Most of ORF1 showed predominant negative selection except for the two regions V1 and V2. V1 showed an elevated dN and an average dS when compared to the ORF1 background but no significant positive selection was detected. The V2 region of ORF1 showed an elevated dN and a low dS when compared to the ORF1 background with an average dN/dS ≈ 3.0 indicative of positive selection. However, the V2 area includes overlapping ORFs, making the dN/dS estimate biased. Joint estimates of the selection intensity in the different ORFs by a recent method indicated that this region of ORF1 was in fact evolving close to neutrality. This was convergent with previous results showing that introduction of stop codons in this region of ORF1 did not impair plant infection. These data suggest that the elimination of a stop codon caused the overprinting of a novel coding region over the ancestral ORF.



Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Baril M, Barkier-Gingras L (2005) Translation of the F protein of Hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acid Res 33:1474–1486
Belshaw R, Pybus OG, Rambaut A (2007) The evolution of genome compression in RNA viruses. Genome Res 17:1496–1504
Chirico N, Vianelli A, Belshaw R (2010) Why genes overlap in viruses. Proc R Soc B 277(1701):3809–3817
Clover GRG, Pearson MN, Elliot DR, Tang Z, Smales TE, Alexander BJR (2003) Characterization of a strain of Apple stem grooving virus in Actinidia chinensis from China. Plant Pathol 52:371–378
Gibbs A, Keese PK (1995) In search of the origin of viral genes. In: Gibbs A, Calisher HC, Garcia-Arenal F (eds) Molecular basis of virus evolution. Cambridge University Press, Cambridge, pp 79–90
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hein J, Støvlbæk J (1995) A maximum-likelihood approach to analyzing nonoverlapping and overlapping reading frames. J Mol Evol 40:181–189
Hirata H, Lu X, Yamaji Y, Kagiwada S, Ugaki M, Namba S (2003) A single silent substitution in the genome of Apple stem grooving virus causes symptom attenuation. J Gen Virol 84:2579–2583
Hirata H, Yamaji H, Komatsu H, Kagiwada S, Oshima K, Okano Y, Takahashi S, Ugaki M, Namba S (2010) Pseudo-polyprotein translated from the full-length ORF1 of capillovirus is important for pathogenicity, but a truncated ORF1 protein without variable and CP regions is sufficient for replication. Virus Res 152:1–9
Holmes EC (2003) Error thresholds and the constraints to RNA virus evolution. Trends Microbiol 11:543–546
Holmes EC (2009) In: Harvey PH, May RM (eds) The evolution and emergence of RNA viruses. Oxford University Press Inc., New York, pp 114–116
Hughes AL (2007) Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity 99:364–373
Inouye N, Maeda T, Mitsuhata K (1979) Citrus tatter leaf virus isolated from Lily. Ann Phytopathol Soc Jpn 45:712–720
Jelkmann W (1995) Cherry virus A: cDNA cloning of dsRNA, nucleotide sequence analysis and serology reveal a newplant capillovirus in sweet cherry. J Gen Virol 76:2015–2024
Keese PK, Gibbs A (1992) Origins of genes: “big bang” or continuous creation? Proc Natl Acad Sci USA 89:9489–9493
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Korber B (2000) HIV signature and sequence variation analysis. In: Allen Rodrigo G, Learn GH (eds) Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, pp 55–72
Kosakovsky Pond SL, Frost SDW (2005) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533
Krakauer DC (2000) Stability and evolution of overlapping genes. Evolution 54:731–739
Kryazhimskiy S, Plotkin JB (2008) The population genetics of dN/dS. PLoS Genet 4(12):e1000304. doi:10.1371/journal.pgen.1000304
Liebenberg A, Wetzel T, Kappis A, Herdemertens M, Krczal G, Jarausch W (2010) Influence of apple stem grooving virus on Malus sieboldii-derived apple proliferation resistant rootstocks. Proceedings of the 21st international conference on virus and other graft transmissible diseases of fruit crops. Julius-Kühn-Archiv 427:186–188
Lister RM, Bancroft JB, Nadakavukaren MJ (1965) Some sap-transmissible viruses from apple. Phytopathology 55:859–870
Magome H, Yoshikawa N, Takahashi T (1999) Single-strand conformation polymorphism analysis of apple stem grooving capillovirus sequence variants. Phytopathology 89:136–140
Magome H, Yoshikawa N, Takahashi T, Ito T, Miyakawa T (1997) Molecular variability of the genomes of capilloviruses from apple, Japanese pear, European pear, and citrus trees. Phytopathology 87:389–396
Martelli GP, Adams MJ, Kreuze JF, Dolja VV (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu Rev Phytopathol 45:73–100
Martin DP, Williamson C, Posada D (2005) RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262
Melcher U (2000) The ‘30 K’ superfamily of viral movement proteins. J Gen Virol 81:257–266
Moury B, Simon V (2011) dN/dS-based methods detect positive selection linked to trade-offs between different fitness traits in the coat protein of Potato virus Y. Mol Biol Evol 28:2707–2717
Nei M, Gojobori T (1986) Simple method for estimating the numbers of synonymous and nonsynonymous substitutions. Mol Biol Evol 3:418–426
Normark S, Bergstrom S, Edlund T, Grundstrom T, Jaurin B, Lindberg FP, Olsson O (1983) Overlapping genes. Annu Rev Genet 17:499–525
Nozawa M, Suzuki Y, Nei M (2009) Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci USA 106:6700–6705
Ohira K, Namba S, Rozanov M, Kusumi T, Tsuchizaki T (1995) Complete sequence of an infectious full-length cDNA clone of citrus tatter leaf capillovirus: comparative sequence analysis of capillovirus genomes. J Gen Virol 76:2305–2309
Pavesi A (2007) Pattern of nucleotide substitution in the overlapping nonstructural genes of influenza A virus and implication for the genetic diversity of the H5N1 subtype. Gene 402:28–34
Sabath N, Landan G, Graur D (2008) A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS ONE 3(12):e3996. doi:10.1371/journal.pone.0003996
Schierup MH, Hein J (2000) Consequences of recombination on traditional phylogenetic analysis. Genetics 156:879–891
Shim H, Min Y, Hong S, Kwon M, Kini D, Kim H, Choi Y, Lee S, Yang J (2004) Nucleotide sequence of a Korean isolate of apple stem grooving virus associated with black necrotic leaf spot disease on pear (Pyrus pyrifolia). Mol Cells 18:192–199
Suzuki Y, Nei M (2004) False-positive selection identified by ML-based methods: examples from the Sig1 gene of the diatom Thalassiosira weissflogii and the tax gene of a human T-cell lymphotropic virus. Mol Biol Evol 21:914–921
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 40. Mol Biol Evol 24:1596–1599
Tatineni S, Afunian MR, Gowda S, Hilf ME, Bar-Joseph M, Dawson WO (2009a) Characterization of the 5- and 3-terminal subgenomic RNAs produced by a capillovirus: evidence for a CP subgenomic RNA. Virology 385:521–528
Tatineni S, Afunian MR, Hilf ME, Gowda S, Dawson WO, Garnsey SM (2009b) Molecular characterization of Citrus tatter leaf virus historically associated with Meyer Lemon trees: complete genome sequence and development of biologically active in vitro transcripts. Phytopathology 99:423–431
Terauchi H, Magome H, Yoshikawa N, Takahashi T, Inouy N (1997) Construction of an infectious cDNA clone of the apple stem grooving capillovirus (isolate Li-23) genome containing a cauliflower mosaic virus 35S RNA promoter. Ann Phytopathol Soc Jpn 63:432–436
Wassenegger M, Heimes S, Sänger HL (1994) An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J 13:6172–6177
Wyckoff GJ, Malcom CM, Vallender EJ, Lahn BT (2005) A highly unexpected strong correlation between fixation probability of nonsynonymous mutations and mutation rate. Tr Genet 21:381–385
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yang Z, Nielsen R, Goldman N, Petersen AM (2000) Codon substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–444
Yang Z, Wong WSW, Nielsen R (2005) Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol 22:1107–1118
Yokoyama S, Tada T, Zhang H, Britt L (2008) Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA 105:13480–13485
Yoshikawa N, Imaizumi M, Takahashi T, Inouye N (1993) Striking similarities between the nucleotide-sequence and genome organisation of citrus tatter leaf and apple stem grooving capillovirus. J Gen Virol 74:2743–2747
Yoshikawa N, Sasaki E, Kato M, Takahashi T (1992) The nucleotide sequence of apple stem grooving capillovirus genome. Virology 191:98–105
Zhou Y, Holmes EC (2007) Bayesian estimates of the evolutionary rate and age of hepatitis B virus. J Mol Evol 65:197–205
Acknowledgments
Annerie Liebenberg was supported by a grant from the Fonds für die Entwicklung ländlicher Räume in Rheinland-Pfalz (F.E.L.R.). We thank Dr. Fiona Constable, Department of Plant Science, Waite Campus, University of Adelaide, Glen Osmond, South Australia, 5064, Australia, Dr. Margit Laimer, Plant Biotechnology Unit, Institute of Applied Microbiology, University of agricultural sciences, Nussdorffer lande 11, 1190 Vienna, Dr. Mike Bernardy, Pacific Agri-Food Research Centre, Agriculture & Agri-Food Canada, Summerland, British Columbia V0H 1Z0, Dr. Pascal Gentit, Centre Technique Inter professionnel des Fruits et Légumes (CTIFL), 22 rue Bergère, 75009 Paris, France, and Michael Petruschke, Landwirtschaftliches Technologiezentrum Augustenberg—Außenstelle Stuttgart, Rötestrasse 16a, 70197 Stuttgart, Germany, for providing the different isolates of ASGV.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liebenberg, A., Moury, B., Sabath, N. et al. Molecular Evolution of the Genomic RNA of Apple Stem Grooving Capillovirus. J Mol Evol 75, 92–101 (2012). https://doi.org/10.1007/s00239-012-9518-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-012-9518-z