Abstract
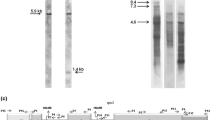
RNA-mediated gene duplication has been proposed to create processed paralogs in the plant mitochondrial genome. A processed paralog may retain signatures left by the maturation process of its RNA precursor, such as intron removal and no need of RNA editing. Whereas it is well documented that an RNA intermediary is involved in the transfer of mitochondrial genes to the nucleus, no direct evidence exists for insertion of processed paralogs in the mitochondria (i.e., processed and un-processed genes have never been found simultaneously in the mitochondrial genome). In this study, we sequenced a region of the mitochondrial gene nad1, and identified a number of taxa were two different copies of the region co-occur in the mitochondria. The two nad1 paralogs differed in their (a) presence or absence of a group II intron, and (b) number of edited sites. Thus, this work provides the first evidence of co-existence of processed paralogs and their precursors within the plant mitochondrial genome. In addition, mapping the presence/absence of the paralogs provides indirect evidence of RNA-mediated gene duplication as an essential process shaping the mitochondrial genome in plants.



Similar content being viewed by others
References
Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29:380–395
Adams KL, Qiu YL, Stoutemyer M, Palmer JD (2002) Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA 99:9905–9912
Allen JF (2003) Why chloroplasts and mitochondria contain genomes. Comp Funct Genomics 4:31–36
Allen JO, Fauron CM, Minx P, Roark L, Oddiraju S, Lin GN, Meyer L, Sun H, Kim K, Wang C, Du F, Xu D, Gibson M, Cifrese J, Clifton SW, Newton KJ (2007) Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177:1173–1192
Bakker FT, Breman F, Merckx V (2006) DNA sequence evolution in fast evolving mitochondrial DNA nad1 exons in Geraniaceae and Plantaginaceae. Taxon 55:887–897
Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD (2004) Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA 101:17747–17752
Bonen L (2008) Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8:26–34
Bonen L, Vogel J (2001) The ins and outs of group II introns. Trends Genet 17:322–331
Bowe LM, dePamphilis CW (1996) Effects of RNA editing and gene processing on phylogenetic reconstruction. Mol Biol Evol 13:1159–1166
Bremer B et al (2009) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161:105–121
Cho Y, Qiu YL, Kuhlman P, Palmer JD (1998) Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci USA 95:14244–14249
Claros MG, Perea J, Shu Y, Samatey FA, Popot J-L, Jacq C (1995) Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. Eur J Biochem 228:762–771
Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, Sun H, Thompson M, Barbazuk WB, Kanuganti S, Tayloe C, Meyer L, Wilson RK, Newton KJ (2004) Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol 136:3486–3503
Cuenca A, Petersen G, Seberg O, Davis J, Stevenson D (2010) Are substitution rates and RNA editing correlated? BMC Evol Biol 10:349. doi:10.1186/1471-2148-10-349
Daley DO, Clifton R, Whelan J (2002) Intracellular gene transfer: reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc Natl Acad Sci USA 99:10510–10515
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–134
Dombrovska O, Qiu YL (2004) Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogenet Evol 32:246–263
Duvall MR, Robinson JW, Mattson JG, Moore A (2008) Phylogenetic analyses of two mitochondrial metabolic genes sampled in parallel from Angiosperms find fundamental interlocus incongruence. Am J Bot 95:871–884
Geiss KT, Abbas GM, Makaroff CA (1994) Intron loss from the NADH dehydrogenase subunit 4 gene of lettuce mitochondrial DNA: evidence for homologous recombination of a cDNA intermediate. Mol Gen Genet 243:97–105
Gindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11:725–736
Gugerli F, Sperisen C, Büchler U, Brunner I, Brodbeck S, Palmer JD, Qiu Y-L (2001) The evolutionary split of Pinaceae from other conifers: evidence from an intron loss and a multigene phylogeny. Mol Phylogenet Evol 21:167–175
Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31:5907–5916
Handa H (2008) Linear plasmids in plant mitochondria: Peaceful coexistences or malicious invasions? Mitochondrion 8:15–25
Knoop V (2004) The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet 46:123–139
Kosakovsky Pond SL, Frost SD, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, Mikami T (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA-Cys (GCA). Nucleic Acids Res 28:2571–2576
Liu SL, Zhuang Y, Zhang P, Adams KL (2009) Comparative analysis of structural diversity and sequence evolution in plant mitochondrial genes transferred to the nucleus. Mol Biol Evol 26:875–891
Maddison WP, Maddison DR (2005) MacClade 4: analysis of phylogeny and character evolution. Version 4.08a
Maddison WP, Maddison DR (2009) Mesquite: a modular system for evolutionary analysis. Version 2.72
Michel F, Ferat JL (1995) Structure and activities of group II introns. Annu Rev Biochem 64:435–461
Moenne A, Begu D, Jordana X (1996) A reverse transcriptase activity in potato mitochondria. Plant Mol Biol 31:365–372
Moritz C, Hillis DM (1990) Molecular systematics: context and controversies. In: Hillis DM, Moritz C (eds) Molecular systematics. Sinauer Ass, Sunderland, MA, pp 1–10
Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 268:434–445
Nugent JM, Palmer JD (1991) RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66:473–481
Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, Miyashita N, Nasuda S, Nakamura C, Mori N, Takumi S, Murata M, Futo S, Tsunewaki K (2005) Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res 33:6235–6250
Parkinson CL, Mower JP, Qiu YL, Shirk AJ, Song K, Young ND, dePamphilis CW, Palmer JD (2005) Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol 5:73
Petersen G, Seberg O, Davis JI, Stevenson DW (2006) RNA editing and phylogenetic reconstruction in two monocot mitochondrial genes. Taxon 55:871–886
Popot J, de Vitry C (1990) On the microassembly of integral membrane proteins. Ann Rev Biophys Biophys Chem 19:369–403
Race H, Herrmann R, Martin W (1999) Why have organelles retained genomes? Trends Genet 15:364–370
Schuster W, Brennicke A (1994) The plant mitochondrial genome: physical structure, information content, RNA Editing, and gene migration to the nucleus. Ann Rev Plant Physiol Plant Mol Biol 45:61–78
Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR (2010) Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics 185:1369–1380
Szmidt AE, Lu MZ, Wang XR (2001) Effects of RNA editing on the coxI evolution and phylogeny reconstruction. Euphytica 118:9–18
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Vanin EF (1985) Processed pseudogenes: characteristics and evolution. Ann Rev Genet 19:253–272
Won H, Renner SS (2003) Horizontal gene transfer from flowering plants to Gnetum. Proc Natl Acad Sci USA 100:10824–10829
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas, Austin
Acknowledgments
The authors thank Charlotte Hansen and Hannah Blossom for skillful assistance in the lab, and the authors acknowledge the DNA Bank at the Royal Botanical Gardens of Kew, Jerrold I. Davis and Dennis W. Stevenson for supplying DNA samples. Financial support was provided by the Danish National Sciences Research Council Grant No. 272-06-0436.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cuenca, A., Petersen, G., Seberg, O. et al. Genes and Processed Paralogs Co-exist in Plant Mitochondria. J Mol Evol 74, 158–169 (2012). https://doi.org/10.1007/s00239-012-9496-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-012-9496-1