Abstract
Tiger nuts (also called earth almonds or chufa) are the stolons of the sedge Cyperus esculentus L. and have been traditionally used as food or as an additive to drinks such as horchata de chufa. Recently, chufa experienced a hype as functional food because of its richness in dietary fiber and as a source of unsaturated fatty acids. Its close relative, nutgrass (Cyperus rotundus L.) has similarly long history of human use, but rather for medical purposes, due to a high content of secondary compounds. Both species are invasive weeds responsible for high yield losses in crop plants, especially in Southeast Asia, Africa, and subtropical America. They are not easily discerned, occur together, and are both traded under similar or identical vernacular names. For the purpose of consumer protection, it is important to discriminate them in commercial samples, which is not trivial by morphological markers and often impossible since they are mostly used in highly processed form. Therefore, a molecular authentication assay was developed using taxonomically authenticated plants as reference material. The plastidic marker psbA-trnH igs (intergenic spacer) and the nuclear marker Internally Transcribed Spacer were used for barcoding. To develop a sequencing-free fingerprinting assay, a single-nucleotide polymorphism in the ITS2 region could be employed to discriminate C. esculentus from its possible adulterant C. rotundus by a tailored Amplificatory Refractory Mutation System PCR. This approach was successful both, in reference to plants, as well as in highly processed commercial products such as tiger nut flour. We show that by a single PCR we can detect surrogation by nutgrass down to a threshold of 10% in tiger nuts as contribution to safeguard product authenticity against possible adulterations in a growing global market.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tiger nut (Cyperus esculentus L.) or the closely related nutgrass or red nut sedge (Cyperus rotundus L.) is a curse and a blessing at the same time—depending on the perspective. As persistent weeds, they cause massive losses in crop yield every year with an effective control strategy still urgently needed [1]. On the other hand, both species have a long history of human use, although the applications differ, while C. esculentus is mainly used for its nutritional value (for reviews see [2, 3]. C. rotundus is less palatable, but richer in secondary compounds and, thus, more used as medicinal plant), for review, see [4].
The subterranean edible stolons of C. esculentus, called tiger nuts or earth almonds, were cultivated already 6000 years ago in the African Mediterranean area [5], and are still important as food or medicine to the present day. The “grains of Al-Aziz” (Hab’ el aziz—the current Egyptian name [6]) are used in different countries: as part of Horchata de chufa (powder of stolons with water and sugar resulting in a refreshing drink) in Spain [7], as addition to local beverages (like Kunnu) in Nigeria and Ghana to increase their nutritional value [8], as general remedy to prevent malnutrition [9], or lately, popularized in Europe in form of complete stolons or flour that can be added to several dishes as functional food. For such applications, only the species C. esculentus is used. This species is morphologically variable with a blurred border between domesticated and wild or even weedy types. The domesticated forms are referred to as var. sativus [10]. More recently, the term var. chufa has been proposed [2]. In addition, several wild and weedy forms with distinct geographic distributions have been described. A thorough taxonomic study by Schippers et al. [11] arrives at four forms, namely C. esculentus var. esculentus, C. esculentus var. leptostachyus (Boeckler), C. esculentus var. hermannii ((Buckley) Britton), and C. esculentus var. macrostachyus (Boeckler). Some authors also delineate C. esculentus var. esculentus as domesticated form from C. esculentus var. sativus [Flora of China, efloras.org]. While the tastes of the domesticated and the wild or weedy forms were shown to be very similar during a comparative study [2], the domesticated var. sativus was more enjoyable because it was found to be less fibrous [12]. A comparison between two accessions of C. esculentus from Nigeria that differed in stolon color [13] found a comparable abundance of essential elements, fatty acids, and carbohydrates [9, 13]. While it is not clear whether these forms belonged to the same type of C. esculentus, the Nigerian study suggests that intraspecific variation apparently does not have big effect on nutritional composition. This is good news for consumer protection, narrowing the target for authentication to the species level (C. esculentus), not calling for the need to differentiate within this species.
However, there is a clear need to draw a line between C. esculentus and C. rotundus, although the contents of bulk nutrients are comparable: Values for carbohydrates in C. esculentus range from around 45% [8] to 59% [13], comparable to the 59% reported for C. rotundus [16]. For lipids, values from 24% [13] to 35% [14] have been found in C. esculentus versus 24% in C. rotundus [16]. For protein content, 4% [15] to 7% [14] in C. esculentus seem to be comparable to the 4.7% reported for C. rotundus [16]. In contrast, the two species differ significantly when the composition of essential oils is considered. The main component of C. esculentus is oleic acid (some 70%), which is comparable to olive oil [8, 15]. In contrast, C. rotundus shows drastically lower contents of oleic acid, only around 5% [17]. Further differences concern palmitic acid that accounted for ~ 15% of the essential oil in C. esculentus [8, 15]), but was not even detectable in C. rotundus [17]. An inverse situation was seen for linoleic acid, which was significantly lower, around 8%–9%, in C. esculentus [8, 15] but very abundant, > 20%, in C. rotundus [17].
Thus, while the two species are similar in nutritional values, they already differ qualitatively in oil composition. These differences are even more pronounced if the secondary compounds are considered that are linked with the rich ethnomedicinal tradition for C. rotundus (for review see [4]). As often in traditional medicinal plants, there are numerous applications, especially in Ayurvedic and other Indian systems of medicine, and the link with the respective active compound is usually unknown (for review, see [18]). However, a few associations have been investigated in more detail, for instance phytoestrogenic activities attributed to β-sitosterols [19] that would explain the use of C. rotundus to cure menstruation-related problems in women [20]. A second group of compounds are phenylpropanoids, such as quercetin or chlorogenic acid that might be responsible for the effect of C. rotundus against inflammatory diseases of the skin [21]. These medically active secondary compounds are not seen in C. esculentus. As long as it is clear that C. rotundus is used in a medical context, where dosage and formulation are controlled by a protocol of use, the use of this species is to be seen as beneficial. The problem arises when this plant becomes isolated from its traditional context of healing and is shifted into a different context, namely, that of C. esculentus, which is used for nutrition. Thus, for the sake of consumer safety, it is important that commercial products from the two species are declared correctly.
This task is far from trivial since the colossal genus Cyperus harbors around 950 species, with newly described species added to this day, such that the taxonomy on Cyperus has remained under continuous revision [22]. This is partially due to the limited set of available morphological traits, rendering taxonomic identification often difficult and ambiguous [23]. Further confusion is added by the fact that many species have been described several times under different names. The number of synonyms listed for certain Cyperus species in the Kew Garden Plant List (now World Flora Online) is overwhelming: for C. esculentus, there are 51 synonyms, for C. rotundus even 76 [theplantlist.org]. Of special interest is the occurrence of a monophyletic subclade of Cyperus with around 760 species that share C4 photosynthesis as synapomorphy and comprise both, C. esculentus and C. rotundus [24], while the species with C3 photosynthesis seem to be paraphyletic. These phylogenetic relationships are supported by the plastidic marker psbA-trnH igs [24], but also by the nuclear markers ETS1f [24], and ITS [25, 26], as well as by complete chloroplast genome data [27].
However, the full diversity of Cyperus is not yet covered by genetic barcoding markers in public databases. For instance, in GenBank, so far only 459 sequences for ITS and 362 for psbA-trnH igs have been deposited (status November 2021), among those certain species are represented with many replicates, others only with one. While a couple of genetic barcoding marker regions are used commonly and have been validated for their universality, ease of sequencing, and discriminative power [28, 29], a region that works equally well in all land plants has not been found yet (and mostly likely never will). Hence, which marker is suitable, needs to be tested individually for the taxon of interest. In plants, nuclear and plastidic genome-based barcodes are used preferentially, while mitochondrial markers, common in animal barcoding, are underrepresented. Among the plastidic markers, psbA-trnH igs has been very powerful, because it is sufficiently universal and often allows discrimination down to the species level [28, 30]. In the monocotyledons, this region is around 350 nucleotides in average, but can reach fragment sizes up to 900 nucleotides [31]. Since the nuclear ITS marker has been widely applied to the genus Cyperus in many previous studies, such that a rich body of information is available, we integrated this marker into our study as well.
For practical applications, DNA fingerprinting methods that do not require sequencing, are preferable. However, methods, such as the use of arbitrary primer pairs, so called RAPD [32] often suffer from drawbacks that impinge on reproducibility. The lack of sequence information has the consequence that annealing temperatures of RAPD primers need to be low and, thus, are prone to target ambiguity [32]. Fingerprinting strategies that use genetic barcoding data can circumvent these specificity issues. The use of Restriction Fragment Length Polymorphism (RFLP) allows to generate species-specific banding pattern by treating amplicons of the respective barcoding marker with an appropriate restriction enzyme. This approach successfully allowed to discriminate between two different plant species of the Myrtaceae family that are both commercialized under the term “Lemon Myrtle” [33], and the same strategy was even able to discern different chemotypes of Holy Basil [34]. As alternative to RFLP, single-nucleotide Polymorphisms (SNPs) can be used to design a duplex PCR with the addition of a diagnostic primer that is tailored such that it will bind only in one species, but not in the other. This strategy, termed Amplification Refractory Mutation System (ARMS), provides the advantage that the full-length amplicon of the barcoding marker serves as a positive control for the success of the PCR as such [35]. In fact, the ARMS method was successfully applied for the identification and authentication of Goji fruits [30], or detecting adulterations of Bamboo Tea [36].
In the current study, we addressed the problem to discriminate C. esculentus and C. rotundus in commercial samples. We started off by (1) constructing a robust framework of authenticated reference plants for molecular barcoding, then (2) implemented the sequences generated from this set of reference plants into a phylogenetic context, which finally (3) allowed applying this context to authenticate the declared identity of commercial samples. Finally, we developed and validated a robust and convenient ARMS PCR-based assay to confirm the presence of C. esculentus and to detect possible adulteration by C. rotundus in processed commercial products.
Materials and methods
Plant material
A collection of reference plants for Cyperus was assembled in the Botanical Garden of the KIT (Table 1) and is maintained as living voucher specimens. Commercial products of tiger nut (traded as dried or powdered stolons) were sampled from supermarkets and online stores (Table 2).
The correct identity of the reference plants is crucial for any authentication approach and, therefore, all reference plants were taxonomically determined using published taxonomic keys [37], Flora of China, http://www.efloras.org), as well as the virtual herbarium of Valdosta State University (https://herb.valdosta.edu). This allowed to verify, in some cases also to revise, the declared identity of these plants [38]. As potential surrogates, almond (Prunus dulcis, Rosaceae), which is often traded in mixtures with tiger nut and in Germany shares the vernacular name (Mandel versus Erdmandel), as well as Oenanthe silaifolia (Apiaceae) forming edible, radish-sized stolons of nutty taste, thus, resembling tiger nut, were included into the study as well, even though they belong to other plant families and are not even monocots.
DNA extraction
DNA from fresh leaves of Cyperus reference plants (using 50 mg of starting material) was isolated using the Invisorb R Spin Plant Mini Kit (Stratec Biomedical AG), following the instructions of the manufacturer.
The processed commercial products were not amenable to DNA extraction using this kit; hence, those samples were extracted using cetyl tri-methyl ammonium bromide (CTAB) according to [39]) with some modifications given in [33]. In brief, the leaf tissue was shock-frozen in liquid nitrogen, and small aliquots (100 mg) were ground using a TissueLyzer (Qiagen, Hilden, Germany). The resulting powder was mixed with 900 µL of boiled extraction buffer (1.5% w/v CTAB) containing 10 µL/mL β-mercaptoethanol, and incubated for one hour at 65 °C. Subsequently, the samples were mixed with 630 µL of chloroform/isoamylalcohol (24:1), shaken horizontally for 15 min, and then spun down for 10 min (17,000g) at room temperature. The upper aqueous phase (which contains the DNA) was transferred into a fresh 2-mL reaction tube, and the DNA was then precipitated with 2/3 v/v of ice-cold isopropanol and subsequently collected by centrifugation (10 min, 17,000g). The sediment was washed with 1 mL 70% EtOH, dried in a vacuum centrifuge for 15 min, and finally dissolved in 50 µL nuclease-free H2O (containing 5 µg RNAse A). Concentration and purity of all DNA samples were determined spectrophotometrically (NanoDrop ND-100, Peqlab).
PCR/gel electrophoresis
As barcoding marker regions, the plastidic psbA-trnH igs and the nuclear Internally Transcribed Spacer (ITS) were amplified in a 30 µL reaction volume containing 20.4 µL nuclease-free water (Lonza, Biozym), 3 µL tenfold Thermopol Buffer (500 mM KCl, 100 mM Tris–HCl and 15 mM MgCl2), 3 µL bovine serum albumin (10 mg/mL), 0.6 µL dNTPs (200 µM, New England Biolabs), 0.6 µL of forward and reverse primer (200 nM, see primer list in Table 3), 0.3 µL of Taq polymerase (5 Units, New England Biolabs), and 1.5 µL of the extracted DNA as template (50 ng/µL).
The psbA-trnH igs region was amplified by initial denaturation at 95 °C for 2 min, followed by 35 cycles with denaturation at 94 °C for 1 min, annealing at 56 °C for 30 s, synthesis at 68 °C for 45 s, and a terminal extension at 68 °C for 5 min prior to storage at 10 °C. The ITS region was amplified using a modified protocol [26] starting with a denaturation at 95 °C for 2 min, followed by nine cycles with denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min (decreasing by 0.7 °C during each cycle), and synthesis at 68 °C for 1 min. This was continued by 19 cycles with denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and synthesis 68 °C for 1 min, followed by a terminal at 68 °C for 7 min, and storage at 4 °C.
The amplicons were evaluated by agarose gel electrophoresis (NEEO ultra-quality agarose, Carl Roth, Karlsruhe, Germany), visualizing the DNA bands by Midori Green (NIPPON Genetics Europe) and blue light excitation (Safe Imager, Invitrogen). The fragment sizes of the amplicons were determined by a comparison with a 100-bp size standard (New England Biolabs). The amplified DNA was purified using the MSB Spin PCRapace kit (Stratec). Sequencing was outsourced to Macrogen Europe (the Netherlands) and Eurofins (Germany). The quality of the obtained sequences was examined using the software FinchTV Version 1.4.01.
Phylogenetic analysis
For the analysis and trimming of the sequences, the MEGA7 software (Version 7.0.14) was employed [40]. A multiple sequence alignment was constructed using the MUSCLE algorithm of MEGA7, and the evolutionary relationships were inferred by using the Neighbor-joining algorithm with a bootstrap value that was based on 1000 replicates [41, 42]. The species Prunus dulcis (almond) and Oenanthe silaifolia served as outgroups to root the phylogenetic tree and to contextualize the genus Cyperus.
Amplified refractory mutation system (ARMS)
Based on single-nucleotide polymorphisms in the ITS2 sequences that were unique for either C. esculentus or C. rotundus, diagnostic primers were designed to trace either of these species. Primer design was performed as described in [30].
For the ARMS diagnostics, the universal ITS2 primers S2F and Ser26 (Table 3) were supplemented by one of those diagnostic primers, respectively, to enable a duplex PCR that should result in the amplification of two fragments for the species of interest, while the alternative species should yield only one band. This approach was also used to verify the identity of the commercial tiger nut products. As to trace tiger nut (C. esculentus), the species-specific ITS_CE3_fw primer was used, for detection of the surrogate, nut grass (C. rotundus), the specific ITS_CR1_fw primer was added instead. The PCR protocol and the reaction mix were as described above, except for the addition of 0.2 µL of the respective diagnostic primers in addition to the primers amplifying the entire ITS fragment see above.
Results
Stolon morphology in reference plants and commercial products is variable
The stolons of Cyperus esculentus varied in size and form, which can be seen in the reference plants as well as in the commercial products. Within the available reference plants, three types were found: round stolons of pale yellow color as in ID9378; crescent-shaped stolons of pale yellow color as in ID3703; and crescent-shaped stolons of dark color as in ID9380 (Fig. 1). The stolons of commercial products (as judged from those with intact organs that had not been processed to a powder) resembled the round, pale yellow type exemplified by ID9378, but were significantly larger. The stolons of ID3704 (Cyperus rotundus) were round as well, but of brown color.
The psbA-trnH igs marker is long in Cyperus and shows a tandem repeat specific for C. esculentus
The length of the psbA-trnH igs sequences in the genus Cyperus ranges from 915 base pairs (after trimming the primer sequences) in the four Cyperus esculentus accessions, to slightly shorter fragments in C3 plants with 875 base pairs in Cyperus diffusus to 891 base pairs in Cyperus eragrostis. The fragments from the two outgroups were conspicuously shorter, with 325 base pairs in almond (Prunus dulcis) and 223 base pairs in Oenanthe silaifolia. The multiple sequence alignment of the Cyperus accessions including the commercial products had a complete length of 1008 nucleotides with a low G/C content of 23.9% in average. The alignment revealed a tandem repeat which was unique for C. esculentus and tiger nut products (Supplementary Figure 1). For all other Cyperus species that were part of this study, this motif, of 14 base pairs in length, occurred only once, while in C. esculentus it was doubled to 28 nucleotides in total, which is one reason for the slightly enhanced length of the C. esculentus sequences. This sequence pattern can be employed to unequivocally discriminate the earth almond from its close relatives, like C. rotundus or Cyperus iria.
The psbA-trnH igs marker separates C3 and C4 Cyperus and confirms commercial tiger nut products
The phylogenetic tree inferred from the psbA-trnH igs dataset reflected the divergence between C3 and C4 Cyperus and placed the surrogate species almond (Rosaceae) as well Oenanthe silaifolia (Apiaceae) as outgroups (Fig. 2). Within the C4 clade, the accessions from Cyperus esculentus reference plants and the six tested commercial products were clearly separated from a sister clade including, among others, C. rotundus and C. iria. Even though the commercial products were partially heavily processed, and the fragment length, with around 1000 base pairs, was fairly large, the fragments could be amplified and sequenced readily. Since all six commercial products not only cluster with the four reference plants of C. esculentus but show the unique 14-bp tandem repeat, they were found to be true tiger nut. Moreover, no sequence ambiguities were observed in the products that could point to possible admixtures. Nevertheless, for the design of a sequencing-free identification assay using ARMS, this repeat is not suited since it almost completely consists of the bases A and T, such that the introduction of destabilizing bases, a central feature of the ARMS strategy, is not possible.
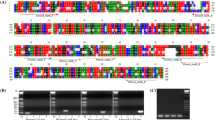
Phylogenetic tree of the plastidic psbA-trnH igs comparing selected Cyperus reference plants and commercial products of tiger nut. The evolutionary history was inferred using the Neighbor-Joining method [42]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [41]. Oenanthe silaifolia and Prunus dulcis form the outgroup. The reference plants of Cyperus are labeled by colored squares, e.g., Cyperus esculentus (green), Cyperus rotundus (red). The evaluated tiger nut commercial products are labeled by gray squares
The Internal Transcribed Spacer is informative and allows for identification of Cyperus
The barcoding marker region Internal Transcribed Spacer could be amplified and sequenced for all reference plants. However, this was not fully successful for the commercial products. In fact, only for one commercial product the complete ITS region (comprising ITS1, 5.8S rDNA, and ITS2) could be amplified and sequenced successfully. For two other products, only the ITS2 region could be sequenced.
The ITS sequences were longer for the C3 species such as C. diffusus, C. eragrostis, and C. involucratus with 599, 594, and 593 base pairs after trimming the primer recognition sites, respectively. For the C4 plants, C. esculentus (550) and C. iria (563), they were shorter. The ITS2 fragments recovered from the commercial products 1 and 5, declared as tiger nut, spanned 288 base pairs for both products. Also those partial sequences were included into the dataset that was used to infer the phylogenetic tree. The length of the two outgroups Prunus dulcis (613 base pairs) and O. silaifolia (600 base pairs) is similar to the Cyperus accessions, however, as dicots due to their large evolutionary distance from the monocot Cyperus, the sequences were, not surprisingly, quite deviant. For the four C. esculentus accessions, significant intraspecific differences were detected (12 parsimonious informative sites in 550 base pairs). However, there are still diagnostic SNPs common to all these accessions that differ from closely related Cyperus species, especially C. rotundus. Two SNPs were exclusively found in C. esculentus over all tested accessions. Thus, these SNPs qualified as diagnostic trait against all other Cyperus species tested in this study. The dataset of the multiple sequence alignment used for inferring the phylogenetic relationship comprises 672 base pairs. The resulting tree places the Rosaceae species P. dulcis and the Apiaceae species O. silaifolia as outgroups (See Fig. 3), and clearly divides Cyperus into a C3 clade from a C4 clade. However, the position of some species within the C4 clade differed from the topology based on the psbA-trnH igs marker. For instance, C. iria was now a sister to C. esculentus, while for the psbA-trnH igs, it had been C. rotundus that emerged as sister. Furthermore, the positioning of C. strigosus differed significantly. This species had been located with the C. iria accessions in the psbA-trnH igs tree but is now basal to the entire C4 clade. Both sequences had been generated from the same DNA sample ruling out mislabelling of samples. Moreover, both psbA-trnH igs and ITS had been assigned as C. strigosus during a BLAST search in GenBank.
Phylogenetic tree of the nuclear ITS region of selected Cyperus reference plants and commercial products of tiger nut. The evolutionary history was inferred using the Neighbor-joining method [42]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [41]. Oenanthe silaifolia and Prunus dulcis form the outgroup. The reference plants of Cyperus are labeled by colored squares, e.g., Cyperus esculentus (green), Cyperus rotundus (red). The evaluated tiger nut commercial products are labeled by gray squares
The varying topologies using nuclear or plastidic markers in the context of Cyperus should be kept in mind when constructing phylogenetic trees and infer the evolutionary relationships of this genus. While all reference plants of Cyperus esculentus as well as the three sequenced commercial products cluster together, despite the presence of intraspecific variations, the overall topology inferred for Cyperus depends on the type of donor genome (plastidic versus nuclear).
Using species-specific signatures for sequencing-free discrimination of closely related Cyperus species
Although there were some difficulties with amplifying and sequencing, the ITS region was more suitable for the ARMS approach mainly because the GC content was higher as compared to psbA-trnH igs, which facilitates tailoring diagnostic primers. One of the diagnostic nucleotide substitutions in the ITS2 region was used to design species-specific primers. In Cyperus esculentus, the ITS2 fragment (including the complete ITS2 region and parts of the 5.8S and 28S rDNA) using the primers listed in Table 2 for amplification has a length of 437 base pairs (including fw and rv primers), whereas the fragment in C. rotundus is slightly shorter with 425 base pairs. The ITS2 fragment in C3 species of the genus Cyperus is slightly larger with 466 base pairs. The decisive SNP (a Guanine) that is diagnostic for C. esculentus is positioned at site 164, resulting (together with the length of the primer) in a second (diagnostic) fragment with a length of 294 base pairs, which can be clearly discerned from the full-length amplicon. In fact, a duplex PCR including primers flanking the ITS2 region along with the diagnostic primer differentially annealing with the species-specific SNP, yielded a second diagnostic band (additionally to the ITS2 fragment) for all four reference plants of Cyperus esculentus (the specific signal is shown for two individuals in Fig. 4b). This diagnostic band was absent in all other species of Cyperus that were part of this study, including the closely related species C. rotundus and C. iria. In the commercial products CP1, CP2, CP3, CP5, CP8 and CP9, the second band could be amplified as well. In contrast, commercial product CP4 lacks this band, even though the full-length ITS2 amplicon is present demonstrating that the PCR had been successful. While this showed that product CP4 is very unlikely to be C. esculentus, it does not tell us, what other species is in the product. We developed, therefore, a complementary assay, using the CR1 primer that is diagnostic for C. rotundus and, in fact, generates a second band in C. rotundus reference plants (See Fig. 4c). This primer did not amplify such a diagnostic band, neither in the reference plants of C. esculentus, nor in the commercial products that had been verified as C. esculentus, further supporting that the closely related C. rotundus was absent from those commercial products. However, sample CP4 that had turned out to be not C. esculentus, showed the second band with primer CR1, which marks the presence of C. rotundus. For both diagnostic primers, an unexpected additional side band was generated at about 1000 bp, that are, however, only amplified in the desired species providing an even clearer signal for C. esculentus or C. rotundus, respectively.
A Schematic illustration of the internal transcribed spacer (ITS) region and orientation of primers used in this study. The decisive nucleotide substitutions were detected in the ITS2 region of Cyperus and sequence cutouts of Cyperus esculentus and Cyperus rotundus as well as the primer sequences are highlighted. B ITS2-based agarose electrophoresis of ARMS amplicons from C. esculentus (green), C. rotundus (red) references and four outgroups (blue, including C. iria, C. strigosus and Oenanthe and Prunus). PCR-result using this diagnostic test including all Cyperus species that were part of this study are shown in Supplementary Figure 2. Commercial products are labeled in gray. The CE3_fw primer that targets C. esculentus was used, see (A). A unique signal in C. esculentus individuals as well as in six of seven commercial products could be detected. C ITS2-based agarose electrophoresis of ARMS amplicons from C. esculentus (green), C. rotundus (red) references and four outgroups (blue, including C. iria, C. strigosus and Oenanthe and Prunus). Commercial products are labeled in gray. The CR1_fw primer that targets C. rotundus was used, see (A). A unique signal in C. rotundus individuals as well as in one of the commercial products could be detected. D AGE of CE3_fw and CR1_fw using reference plants and artificial DNA mixtures of C. esculentus/C. rotundus (using 90/10, 70/30, 50/50, 30/70, and 10/90 mixtures) in order to test the sensitivity of the approach with respect to the detection of possible adulterations. Independent of concentration, adulteration by the other species could be demonstrated
To determine the detection limit of these ARMS assays, we prepared artificial mixtures of C. esculentus and C. rotundus DNA, thus simulating surrogations in commercial products that would be otherwise hard to identify, especially in powders. An artificial mixture of 90% C. esculentus and 10% C. rotundus DNA still showed the second diagnostic band using the CR1 primer, which means that admixtures of C. rotundus can be picked up down to 10% (See Fig. 4d).
Discussion
Globalization has not only an economic, but also a botanical component. Plants and plant products are traded globally, entering novel markets. Wealthy, but aging societies develop an increasing awareness for health, while on the other hand, the pressure for economic performance fuel self-optimisation. Both developments drive a boom for functional foods that are consumed for their presumed effect upon human health in the first place. Many of these functional foods originate from traditional medicinal systems, such as Ayurveda, Unani, or Traditional Chinese Medicine that employ food as tools to prevent disease. When these plants are separated from their traditional cultural context and become traded globally, this can result in confusion, or even consumer deception. The attempt to regulate the flow of these new and exotic products by legislation has not been overly successful as illustrated by the fact that meta-studies on adulteration of plant products arrive at estimates of > 25% of cases, where the declaration conflicts with the actual content of a sample (for review, see [46]).
Adulteration is facilitated by situations, where products are entering new markets, whose demand is increasing dynamically, such that production cannot keep pace. These criteria are met by the tiger nut market. The global import volume for tiger nut in 2021 was estimated to range around 2400 million US dollars with a yearly growth rate of close to 20% [47]. Main importer, with around 690 million US dollars, is still the US, but there, growth seems saturated with a five-year increase of only 3.9%. In contrast, China (559 million US dollars, five-year increase of > 150%) and Germany (229 million US dollars, five-year increase of 62%) emerge as new major importers. Under these circumstances, a controlled regional production would be the best option to control authenticity and avoid adulteration of tiger nut products. However, C. esculentus has been included into a list of 150 species that are banned from being introduced to the European Union based on the assessment of the European Food Safety Authority with the intention to contain the phytopathogenic bacterium Xylella fastidiosa [48]. Thus, the import of tiger nut products to Europe is expected to grow at similar rates over the next years, such that adulteration will become a progressively pertinent issue. In the following, we will discuss, therefore, what factors make tiger nut prone to fall prey to adulteration (both, intentionally and non-intentionally), how the genetic barcoding strategy developed in the current work can contribute to prevent this, where the limitations of our approach are, and how these could be overcome in future.
Why Cyperus causes chaos—in crop fields, in taxonomy and in food products
Stolons derive from underground axillaries and develop by swelling in moist and dark atmosphere [49]. This tuberisation process is not only dependent on genetic factors, but also various environmental conditions, such as soil depth. For instance, for potatoes, cool temperatures [50] and low levels of nitrogen [51] promote tuberisation, while tubers develop more slowly, when it is warm, or when nitrogen is not limiting. These environmental conditions act through modulation of phytohormone synthesis and signaling. For tuberisation in potatoes, especially gibberellic acid (the molecular mechanics are reviews in [52, 53]) and jasmonic acid [54, 55] have been found to be relevant.
Thus, the morphological variability of stolons that was observed for C. esculentus in the current study, but also reports from other authors, while certainly also depending on genetic factors [11] is most likely also depending on environmental conditions including abiotic stress responses. For C. rotundus, comparative studies have revealed an influence of flooding [56] and differences in rainfall [57] as modifiers of tuberisation. This morphological diversity of stolons seems not to be accompanied by major differences of composition, at least, if bulk compounds are considered [9, 13]. On the other hand, given this variability of stolon morphology, this trait does not qualify as reliable taxonomic marker to delineate the two species C. esculentus and C. rotundus (see Fig. 1), which might be the reason, why determination keys do not use it as discriminative marker. Anyway, most commercial products are not traded as intact stolons, but often come in processed form, for instance, as flour, such that the anyway shaky identification by stolon morphology is rendered impossible.
Since both species also spread as invasive weeds, the prospect to use a pertinent weed as highly priced functional foods is certainly too tempting for farmers in developing countries to be missed, and the rapid growth of chufa markets promote this tendency even further. A further factor might be that vernacular taxonomy is often ambiguous, as exemplified by the use of the species in Ayurveda. C. rotundus plays an important role in Ayurvedic medicine and is designated, among other names, as musta and delineated in the Vedic scripts from jala musta, representing C. esculentus [58], www.easyayurveda.com] [59]. However, in current use, this fine line is often ignored and jala musta just described as a “variant” of musta [60]. It does not need a wild imagination to conceive that, for instance, a German importer might easily fall prey to these pitfalls of traditional nomenclature with the risk of non-intentional adulteration.
psbA-trnH igs and ITS barcoding regions separate C. esculentus and C. rotundus
The choice for the “best” genetic barcoding marker depends on the species or genus of interest. Many different marker regions have been proposed in theory to be the most suitable. However, in practice, the trial of several barcodes is unavoidable. For instance, the “most promising plastid DNA barcode of land plants [61]”, the ycf1b region, could not be amplified at all in the Cyperus accessions of this study.
The plastidic psbA-trnH igs displayed a complete length of around 1000 base pairs in chufa, which is remarkably large for this barcoding region, considering the average length for monocotyledons being 357 base pairs, with an estimated maximum of 905 base pairs [31]. The probability of SNPs that can be utilized for molecular authentication through RFLP or ARMS enhances with length; however, low G/C-content in chufa psbA-trnH igs limits especially the ARMS approach since the design of viable primers depends among other factors on a sufficient G/C content. While longer barcoding markers have a higher chance to find informative substitutions, the enhanced length of is also a drawback since during the processing of commercial products the DNA can be degraded to such a degree, that a fragment with a size of 1000 base pairs cannot be amplified.
The phylogenetic tree based on the plastidic marker psbA-trnH igs gives an insight on the maternal evolution of a small fraction from the huge Cyperus genus (See Fig. 2) and helps putting the species of interest C. esculentus and C. rotundus into a phylogenetic context. The clear separation of the Cyperus C3 and C4 clades has been described earlier using the plastidic psbA-trnH igs [24] and is consistent with previous studies that had shown that this genus is not monophyletic (see [25, 26]). Within the C4 clade, C. esculentus, C. rotundus, and Cyperus iria are close, but clearly separated. The topology for the nuclear ITS region was fairly close although we encountered some difficulties in addressing the commercial products because either amplification or the sequencing failed. This does not come as a surprise, since the nuclear genome is only present in two copies, while the plastidic genome is more abundant in cells. The more abundant template can, therefore, explain the higher success rate for the psbA-trnH igs marker. Most importantly, irrespective of the marker, the commercial products clustered together with C. esculentus.
Although the overall patterns were close, the plastidic and the nuclear marker produced one significant difference in topology. While C. iria was clearly placed with C. rotundus for the plastidic psbA-trnH igs region (see Fig. 2), it is as clearly close to C. esculentus, when the ITS marker is considered (see Fig. 3). Since plastids are, in most cases, maternally inherited, while nuclear genomes are symmetrically passed on, a straightforward explanation might be that C. iria derived from a hybridisation event, where a species close to C. esculentus, acted as pollen recipient. Alternatively, more exotic mechanisms, such as chloroplast capture might play a role [62], which have been demonstrated in Quercus [63] or Vigna [64]. Though chloroplast capture has not been reported in Cyperus, the readiness for asexual reproduction through stolons might favor the occurrence of such an event. Generally, one needs to keep in mind that radiation of Cyperus was a recent event. For instance, the geographically different subspecies of C. esculentus have diverged approximately only 5.1 million years ago [65]. Thus, alleles shared between different species might result in incomplete lineage sorting as it has been shown in other monocot groups such as the subtribe Hippeastrinae from the Amarayllidaceae family [66] or genus Allium from the same family [67]. Therefore, more loci, both plastidic and nuclear, would be needed to address a potential discrepancy of plastidic and nuclear phylogenies. However, the scope of the current study was not to address the evolution of the complex genus Cyperus, but, quite pragmatically, to use the detected sequence polymorphisms to develop a robust and sequencing-free assay that allows to distinguish C. esculentus and C. rotundus.
Beyond phylogeny: sequencing-free authentication of C. esculentus
The scope of the current study was to develop a robust assay that allows to discern C. esculentus and C. rotundus by a sequencing-free ARMS strategy. Compared to RFLP as alternative method that can amplify discriminative SNPs into a diagnostic banding pattern [35], the ARMS approach does not require a second step (restriction) after the PCR, and also includes a positive control in form of the full-length amplicon as readout of the successful PCR [68, 69]. Which barcoding marker can be used depends on the species—while the psbA-trnH igs marker was successful to authenticate Lycium barbarum, traded as ‘Goji’ [30] or the Peruvian Amaranth (Amaranthus caudatus) known under the vernacular name of kiwicha [70], this marker was not amenable for ARMS in the current study. Instead, the ITS2 region that was successfully employed in the current study, did not work in case of Amaranth, also due to un-favorable AT content. Whether a marker can be used for the ARMS strategy is, thus, not only depending on its resolving powers, but also on its base composition. It is virtually impossible to propose a priori a barcode that would allow for ARMS in all species, this can only be decided after aligning the sequences from the target species and its surrogates.
In the current study, we could identify a diagnostic region in the ITS2 marker and develop two complementary ARMS assays—one produced the diagnostic side band, when the specimen contained C. esculentus, the other, when C. rotundus was present. The assay worked also for processed material, such as chufa flour, underlining the sensitivity of this assay and its importance for consumer safety, and detected admixtures down to 10%. The complementarity is needed to allow for different applications. To verify that a given sample contains tiger nut, the assay is needed, where the side band reports C. esculentus. However, in order to verify that this sample is not surrogated by C. rotundus, the second assay is useful, because, here, a second band inevitably reports that the sample has been adulterated.
Limitation and potential of the assay
Any identification method, no matter how broad in scale, is only as good as the reference material is reliable. This is also valid for DNA-based approaches. However, in addition to authenticity of the references also, the quality of the DNA is crucial [71]. In commercial samples, processing including high temperature or low pH (reviewed in [72] will affect the quality of the DNA and thereby all downstream molecular approaches. Degraded DNA may not deliver a functional template for amplification of long (> 500 bp) barcoding marker fragments, such as rbcLa, ycf1b or ITS. This limitation can be circumvented by choosing shorter fragments spanning the informative region. For instance, the nuclear ITS region contains the central, highly conserved 5.8S region that can be utilized for amplification of the shorter ITS1 and ITS2 fragments. For degraded DNA were templates of > 200 bp are typically rare, it is even possible to shorten the amplicon length to fragments of even only 50–150 bp around the diagnostically relevant polymorphism. The resulting amplicons are still readily separated on an agarose gel. The drawback of such tailored approaches is that not only the ARMS primers, but also the flanking primers need to be designed for the specific assay, and cannot readily be transferred to assays for other species [73, 74].
Recently, hyperspectral fingerprints in the NIR range have been proposed for the differentiation of plants. For instance, weeds in cereal fields can be detected by this approach (knapweed and weedy barley in wheat: Hermann et al. 2013; Weedy oat in wheat [74]). Even on the subspecies level, varieties of maize could be discriminated by this approach [75]. A similar strategy allowed to discriminate C. esculentus (here, in a weed context) from other Cyperaceae, such as Carex hirta L. [76]. To what extent, hyperspectral fingerprints can be used to verify tubers or processed products from C. esculentus against C. rotundus remains unclear. The spectra are highly depending on the reflective properties of the specimen, such that the approach requires extensive calibration and standardization [78], which appears to be a serious limitation compared to a DNA-based approach that is independent of environmental conditions.
Our assay is targeted, it discriminates a specified species against surrogation by a different species. This targeted approach becomes limited, when the commercial product is a mixture of different plant species, because the resulting patterns become complex and difficult to interpret, as demonstrated for mixed samples with Holy Basil [77]. Here, we are currently testing a combination of DNA barcoding and Next-Generation Sequencing that would allow to get statistical information about the abundance of different amplicons.
Conclusions
The rapidly growing markets for botanicals with functional properties bear the possibility for adulterations (accidently or on purpose). We have developed a convenient, sequencing-free tool to authenticate C. esculentus against the closely related C. rotundus that is used in a different context, as medicinal plant. Such robust and reliable assays are needed to protect consumer safety against the challenges of botanical globalization.
Data availability
The sequencing data have been deposited in ncbi database.
References
Holm LG, Plucknett DL, Pancho JV, Herberger JP (1977) The world’s worst weeds. Distribution and biology. University Press of Hawaii
De Vries FT (1991) Chufa (Cyperus esculentus, Cyperaceae): a weedy cultivar or a cultivated weed? Econ Bot 45:27–37
Zhang S, Li P, Wei Z, Cheng Y, Liu J, Yang Y, Wang Y, Mu Z (2022) Cyperus (Cyperus esculentus L.): a review of its compositions, medical efficacy, antibacterial activity and allelopathic. Plants 11:1127
Taheri Y, Herrera-Bravo J, Huala L, Salazar LA, Sharifi-Rad J, Akram M, Cho WC (2021) Cyperus spp.: a review on phytochemical composition, biological activity, and health-promoting effects. Oxidat Med Cell Longevity
Zohary D, Hopf M (1988) Domestication of plants in the old world. Clarendon, pp 415–421
Bayer G (1957) Das Mandelzypergras (Cyperus esculentus L.). Planta Med 5:84–94
Sánchez-Zapata E, Fernández-López J, Angel Pérez-Alvarez J (2012) Tiger nut (Cyperus esculentus) commercialization: health aspects, composition, properties, and food applications. Comprehens Rev Food Sci Food Safety 11:366–377
Sabah MS, Shaker M, Moursy I (2019) Nutritional value of Tiger nut (Cyperus esculentus l.) Tubers and its products. Environ Sci 14:301–318
Imam TS, Aliyu FG, Umar HF (2013) Preliminary phytochemical screening, elemental and proximate composition of two varieties of Cyperus esculentus (Tiger nut). Nigerian J Basic Appl Sci 21:247–251
Böckler J (1870) Die Cyperaceeen des königlichen Herbariums zu Berlin. Linnaea 36:287–291
Schippers P, Ter Borg SJ, Bos JJ (1995) A revision of the infraspecific taxonomy of Cyperus esculentus (Yellow Nutsedge) with an experimentally evaluated character set. Syst Bot 20:461–481
http://www.efloras.org/florataxon.aspx?flora_id=2&taxon_id=109010, Accessed 30 August 2022
Ihenetu SC, Ibe FC, Inyamah PC (2021) Comparative study of the properties of yellow and brown Cyperus esculentus L. World News Natl Sci 35:25–37
Oladele AK, Aina JO (2007) Chemical composition and functional properties of flour produced from two varieties of tigernut (Cyperus esculentus). Afr J Biotech 6:2473–2476
Adel AAM, Awad AM, Mohamed HH, Iryna S (2015) Chemical composition, physicochemical properties and fatty acid profile of Tiger Nut (Cyperus esculentus L) seed oil as affected by different preparation methods. Int Food Res J 22:1931–1938
Ikon GM, Etang UE, Udoiko EM, Ohagim IP (2020) Evaluation of phytochemical contents, proximate nutritional composition and antimicrobial activity of the leaves and rhizome extracts of Cyperus rotundus Linn. Uyo, Akwa Ibom State, Nigeria. South Asian J Res Microbiol 7:1–11
El-Wakil EA, Morsi EA, Abel-Hady H (2019) Phytochemical screening, antimicrobial evaluation and GC-MS Analysis of Cyperus rotundus. World J Pharm Pharmaceut Sci 8:129–139
Dhar P, Dhar DG, Rawat AKS, Srivastava S (2017) Medicinal chemistry and biological potential of Cyperus rotundus Linn.: an overview to discover elite chemotype (s) for industrial use. Ind Crops Prod 108:232–247
Sonwa MM, König WA (2001) Chemical study of the essential oil of Cyperus rotundus. Phytochemistry 58:799–810
Bora D, Mehmud S, Das KK, Medhi H (2016) Report on folklore medicinal plants used for female health care in Assam (India). Int J Herb Med 4:4–13
Rocha FG, de Mello Brandenburg M, Pawloski PL, da Silva Soley B, Costa SCA, Meinerz CC, Cabrini DA et al (2020) Preclinical study of the topical anti-inflammatory activity of Cyperus rotundus L. extract (Cyperaceae) in models of skin inflammation. J Ethnopharmacol 254:112709
Ribeiro ARDO, Pereira-Silva L, Vieira JPS, Larridon I, Ribeiro VS, Felitto G, Alves M et al (2021) Cyperus prophyllatus: an endangered aquatic new species of Cyperus L. (Cyperaceae) with a exceptional spikelet disarticulation pattern among about 950 species, including molecular phylogenetic, anatomical and (micro) morphological data. PLoS ONE 16:e0249737
http://www.theplantlist.org. Accessed 09 August 2021
Larridon I, Bauters K, Reynders M, Huygh W, Muasya AM, Simpson DA, Goetghebeur P (2013) Towards a new classification of the giant paraphyletic genus Cyperus (Cyperaceae): phylogenetic relationships and generic delimitation in C4 Cyperus. Bot J Linn Soc 172:106–126
Reid CS, Doyle VP, Carter JR, Vargas-Rodriguez Y, Urbatsch LE (2017) Molecular systematics of targeted flat sedges (Cyperus, Cyperaceae) of the Americas. Plant Ecol Evolut 150:343–357
Reid CS, Carter R, Urbatsch LE (2014) Phylogenetic insights into New World Cyperus (Cyperaceae) using nuclear ITS sequences. Brittonia 66:292–305
Yang LL, Niu JQ, Tang WW (2021) The complete chloroplast genome of pioneering plant Cyperus iria L. (Cyperaceae) in ecological restoration. Mitochondrial DNA Part B 6:1335–1336
Kress WJ et al (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci 102:8369–8374
CBOL Plant Working Group 1 (2009) A DNA barcode for land plants. Proc Natl Acad Sci 106:12794–12797
Wetters S, Horn T, Nick P (2018) Goji who? Morphological and DNA based authentication of a “superfood.” Front Plant Sci 9:1859
Pang X et al (2012) Utility of the trnH–psbA intergenic spacer region and its combinations as plant DNA barcodes: a meta-analysis. PLoS ONE 7:e48833
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Horn T, Barth A, Rühle M, Häser A, Jürges G, Nick P (2012) Molecular diagnostics of Lemon Myrtle (Backhousia citriodora versus Leptospermum citratum). Eur Food Res Technol 234:853–861
Jürges G, Sahi V, Rios Rodriguez D, Reich E, Bhamra S, Howard C, Slater A, Nick P (2018) Product authenticity versus globalisation—The Tulsi case. PLoS ONE 13:e0207763
Horn T, Völker J, Rühle M, Häser A, Jürges G, Nick P (2014) Genetic authentication by RFLP versus ARMS? The case of Moldavian Dragonhead (Dracocephalum moldavica L.). Eur Food Res Technol 238:93–104
Horn T, Häser A (2016) Bamboo tea: reduction of taxonomic complexity and application of DNA diagnostics based on rbcL and matK sequence data. Peer J 4:e2781
Schmeil O, Fitschen J (2011) Flora Von Deutschland und Angrenzender Länder, vol 95. Quelle Meyer Verlag, Wiebelsheim
The Valdosta State University Virtual Herbarium, https://herb.valdosta.edu/search-specimens.php. Accessed 20 September 2022
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evolut 33:1870–1874
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Gu W, Song J, Cao Y, Sun Q, Yao H, Wu Q, Chao J, Zhou J, Xue W, Duan J (2013) Application of the ITS2 region for barcoding medicinal plants of Selaginellaceae in Pteridophyta. PLoS ONE 8:e67818
Sang T, Crawford DJ, Stuessy TF (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot 84:1120–1136
Tate JA, Simpson BB (2003) Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot 28:723–737
Ichim MC, Häser A, Nick P (2020) Microscopic authentication of commercial herbal products in the globalized market: potential and limitations. Front Pharmacol 11:876
Tridge Product Browser (2022) Tiger Nut. https://www.tridge.com/intelligences/tiger-nut/import Accessed 03 November 2022
European Food Safety Authority Panel on Plant Health (2015) Scientific opinion on the risk to plant health posed by Xylella fastidiosa in the EU territory, with the identification and evaluation of risk reduction options. Eur Food Safety Author J 13:3989
Kumar D, Wareing PF (1972) Factors controlling stolon development in the potato plant. New Phytol 71:639–648
Gregory LE (1956) Some factors for tuberization in the potato plant. Am J Bot 43:281–288
Gao Y, Jia L, Hu B, Alva A, Fan M (2014) Potato stolon and tuber growth influenced by nitrogen form. Plant Product Sci 17:138–143
Fernie AR, Willmitzer L (2001) Molecular and biochemical triggers of potato tuber development. Plant Physiol 127:1459–1465
Kondhare KR, Natarajan B, Banerjee AK (2020) Molecular signals that govern tuber development in potato. Int J Dev Biol 64:133–140
Koda Y, Kikuta Y, Tazaki H, Tsujino Y, Sakamura S, Yoshihara T (1991) Potato tuber-inducing activities of jasmonic acid and related compounds. Phytochemistry 30:1435–1438
Pelacho AM, Mingo-Castel AM (1991) Jasmonic acid induces tuberization of potato stolons cultured in vitro. Plant Physiol 97:1253–1255
Fuentes RG, Baltazar AM, Merca FE, Ismail AM, Johnson DE (2010) Morphological and physiological responses of lowland purple nutsedge (Cyperus rotundus L) to flooding. Ann Bot Plants. https://doi.org/10.1093/aobpla/plq010
Tachie-Menson JW, Buah JN, Adu MO, Afutu E (2021) Morphological diversity in purple nutsedge from four agro-ecological zones in Ghana. Heliyon 7:e07661
Jagtap AG, Shirke SS, Phadke AS (2004) Effect of polyherbal formulation on experimental models of inflammatory bowel diseases. J Ethnopharmacol 90:195–204
Jalamusta https://www.easyayurveda.com/2015/01/07/musta-cyperus-rotundus-uses-research-side-effects/. Accessed 16 November 2022
Bhaskar A (2022) Drugs acting on respiratory system—their properties and mode of action according to Ayurveda. World J Pharmaceut Med Res 8:60–69
Dong W, Xu C, Li C, Sun J, Zuo Y, Shi S, Zhou S (2015) ycf1, the most promising plastid DNA barcode of land plants. Scient Rep 5(1):1–5
Tsitrone A, Kirkpatrick M, Levin DA (2003) A model for chloroplast capture. Evolution 57(8):1776–1782
Whittemore AT, Schaal BA (1991) Interspecific gene flow in sympatric oaks. Proc Natl Acad Sci 88(6):2540–2544
Pasquet RS, Feleke Y, Gepts P (2021) Cowpea [Vigna unguiculata (L.) Walp.] maternal lineages, chloroplast captures, and wild cowpea evolution. Genet Resour Crop Evolut 68(7):2799–2812
De Castro O, Gargiulo R, Del Guacchio E, Caputo P, De Luca P (2015) A molecular survey concerning the origin of Cyperus esculentus (Cyperaceae, Poales): two sides of the same coin (weed vs. crop). Annals Botany 115(5):733–745
García N, Folk RA, Meerow AW, Chamala S, Gitzendanner MA, de Oliveira RS, Soltis PS (2017) Deep reticulation and incomplete lineage sorting obscure the diploid phylogeny of rain-lilies and allies (Amaryllidaceae tribe Hippeastreae). Mol Phylogenet Evolut 111:231–247
Gurushidze M, Fritsch RM, Blattner FR (2010) Species-level phylogeny of Allium subgenus Melanocrommyum: incomplete lineage sorting, hybridization and trnF gene duplication. Taxon 59(3):829–840
Ren W, Guo D, Xing G, Yang C, Zhang Y, Yang J, Yang X (2021) Complete chloroplast genome sequence and comparative and phylogenetic analyses of the cultivated Cyperus esculentus. Diversity 13(9):405
Wu R, Yu C, Wu Y (2021) Characterization of the complete plastome of Cyperus rotundus L. (Cyperaceae). Mitochondr DNA Part B 6(1):58–59
Kanbar A, Beisel J, Wetters S, Gutierrez MT, Graeff-Hönninger S, Nick P (2022) A rapid, simple, and reliable assay to authenticate Peruvian kiwicha (A. caudatus) for food applications. Eur Food Res Technol 11:1–19
Wetters S, Sahi V, Brosche L, Häser A, Nick P (2023) Monitoring Indian “Superfood” Moringa oleifera L.—species-specific PCR-fingerprint-based authentication for more consumer safety, under review
Gryson N (2010) Effect of food processing on plant DNA degradation and PCR-based GMO analysis: a review. Anal Bioanal Chem 396(6):2003–2022
Herrmann I, Shapira U, Kinast S, Karnieli A, Bonfil DJ (2013) Ground-level hyperspectral imagery for detecting weeds in wheat fields. Precis Agric 14(6):637–659
López-Granados F, Pena-Barragán JM, Jurado-Expósito M, Francisco-Fernández M, Cao R, Alonso-Betanzos A, Fontenla-Romero O (2008) Multispectral classification of grass weeds and wheat (Triticum durum) using linear and nonparametric functional discriminant analysis and neural networks. Weed Res 48(1):28–37
Chivasa W, Mutanga O, Biradar C (2019) Phenology-based discrimination of maize (Zea mays L.) varieties using multitemporal hyperspectral data. J Appl Remote Sens 13(1):017504
Lauwers M, De Cauwer B, Nuyttens D, Cool SR, Pieters JG (2020) Hyperspectral classification of Cyperus esculentus clones and morphologically similar weeds. Sensors 20(9):2504
Ríos Rodríguez D, Sahi VP, Nick P (2021) Authentication of Holy basil using markers relating to a toxicology-relevant compound. Eur Food Res Technol 247:2485–2497
Tirado SB, St S, Dennis TA, Enders NM (2021) Utilizing spatial variability from hyperspectral imaging to assess variation in maize seedlings. Plant Phenome J 4:e20013
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wetters, S., Häser, A., Ehrlich, T. et al. Tracing tiger nut (C. esculentus L.): functional food from the colossal Cyperus genus. Eur Food Res Technol 250, 225–238 (2024). https://doi.org/10.1007/s00217-023-04382-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-023-04382-y