Abstract
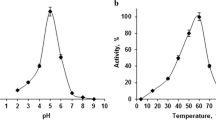
The production of monoglycosylated flavonoids by α-l-rhamnosidases (EC 3.2.1.40) is an interesting development in biocatalysis. Applications of rhamnosidases in industry include removal of bitterness caused by naringin from citrus juices. In the present work, a psychrotolerant bacterial strain with α-l-rhamnosidase activity was isolated. The α-l-rhamnosidase was found to be able to degrade naringin and was purified and characterized. The α-l-rhamnosidase from Brevundimonas sp. Ci19 was able to release both rhamnose and prunin from naringin. The enzyme was partially purified with a performance of 2.7-fold purification. The α-l-rhamnosidase showed an optimum pH between 6.00 and 7.00 with substantial residual activity at pH 5.00 (85.3 %). The optimum temperature was between 20 and 37 °C. The enzyme showed activation in the presence of Ca2+ and Cd2+ ions and at a high ethanol concentration level (10 % v/v). Activity was found for β-d-glucosidase (EC 3.2.1.21) in the partially purified extract, but it was inactive in the acid pH region. This result indicates the potential for inactivation of β-d-glucosidase along with the high level of α-l-rhamnosidase activity necessary for the production of flavonoid glycosides. The α-l-rhamnosidase from Brevundimonas sp. Ci19 showed interesting properties for potential use not only in the citrus juice industry but also in winemaking.





Similar content being viewed by others
References
Vila-Real H, Alfaia AJ, Bronze MR, Calado ART, Ribeiro MHL (2011) Enzymatic synthesis of the flavone glucosides, prunin and isoquercetin, and the aglycones, naringenin and quercetin, with selective α-l-rhamnosidase and β-d-glucosidase activities of naringinase. Enzyme Res. doi:10.4061/2011/692618
Tuberoso CI, Montoro P, Piacente S, Corona G, Deiana M, Dessì MA, Pizza C, Cabras P (2009) Flavonoid characterization and antioxidant activity of hydroalcoholic extracts from Achillea ligustica All. J Pharm Biomed Anal 50(3):440–448
Beekwilder J, Marcozzi D, Vecchi S, de Vos R, Janssen P, Francke C, van Hylckama Vlieg J, Hall RD (2009) Characterization of rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl Environ Microbiol 75(11):3447–3454
Natera Rivas JJ, Batista Zamora AE (2010) El complejo agroindustrial limonero de la provincia de Tucumán (Argentina). Ejemplo de producciones no tradicionales y de desaparición de los pequeños productores. Boletín de la Asociación de Geógrafos Españoles 53:67–88
Cheynier V (2006) In Anderson OM, Markham KR (Eds.) Flavonoids: Chemistry, biochemistry and applications. Taylor & Francis Group, New York: Boca Raton, pp 263–370
Orrillo G, Ledesma P, Delgado O, Spagna G, Breccia J (2007) Cold-active α -l-rhamnosidase from psychrotolerant bacteria isolated from a sub-Antarctic ecosystem. Enzyme Microb Tech 40:236–241
Illanes A, Cauerhff AA, Wilson L, Castro GR (2012) Recent trends in biocatalysis engineering. Bioresource Technol 115:48–57
Yadav V, Yadav PK, Yadav S, Yadav KDS (2010) α -l-Rhamnosidase: a review. Process Biochem 45:1226–1235
Debashish G, Malay S, Barindra S, Joydeep M (2005) Marine enzymes. Adv Biochem Engin/Biotechnol 96:189–218
Fritz I, Strömpl C, Nikitin DI, Lysenko AM, Wolf-Rainer A (2005) Brevundimonas mediterranea sp. nov., a non-stalked species from the Mediterranean Sea. Int J Syst Evol Microb 55:479–486
Alvarenga AE, Pereira EC, Cristóbal HA, Abate CM (2012) In: Amoroso MJ, Benimeli CS, Cuozzo SA (eds.) Actinobacteria: Application in Bioremediation and Production of Industrial Enzymes Tehran, Irán. Science Publishers. p 274–284
Hoffman CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272
Saitou N, Nei M (1987) The neighbor-joining method, a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 15(227):680–685
Ribeiro MH, Manha S, Brito L (2006) The effects of salt and pH stress on the growth rates of persistent strains of Listeria monocytogenes collected from specific ecological niches. Food Res Int 39(7):816–822
Pucci G, Acuña A, Llanes ML, Tiedemann MC, Pucci OH (2009) Diversity of cultivable bacteria from the coast of Caleta Olivia, Patagonia, Argentina. Acta Biol Colomb 14(3):121–134
Tsubouchi T, Shimane Y, Usui K, Shimamura S, Mori K, Hiraki T, Tame A, Uematsu K, Maruyama T, Hatada Y (2012) Brevundimonas abyssalis sp. nov., a dimorphic prosthecate bacterium isolated from deep subseafloor sediment in Japan. Int J Syst Evol Microbiol. doi:10.1099/ijs.0.043364-0
Manzanares P, Orejas M, Ibañez E, Vallés S, Ramón D (2000) Purification and characterization of an α-l-rhamnosidase from Aspergillus nidulans. Lett Appl Microbiol 31:198–202
Qian S, Yu H, Zhang C, Lu M, Wang H, Jin F (2005) Purification and characterization of dioscin-alpha-l-rhamnosidase from pig liver. Chem Pharm Bull 53:911–914
Yanai T, Sato M (2000) Purification and characterization of an α-l-rhamnosidase from Pichia angusta X349. Biosci Biotechnol Bioch 64:2179–2185
Miake F, Satho T, Takesue H, Yanagida F, Kashige N, Watanabe K (2000) Purification and characterization of intracellular α-l-rhamnosidase from Pseudomonas paucimobilis FP2001. Arch Microbiol 173:65–70
Ichinose H, Fujimoto Z, Kaneko S (2013) Characterization of an α-l-rhamnosidase from Streptomyces avermitilis. Biosci Biotechnol Biochem 77(1):213–216
Fujimoto Z, Jackson A, Michikawa M, Maehara T, Momma M, Henrissat B, Gilbert HJ, Kaneko S (2013) The structure of a Streptomyces avermitilis α-l-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement. J Biol Chem 288(17):12376–12385
Gallego MV, Pinaga F, Ramon D, Valles S (2001) Purification and characterization of an α-l-rhamnosidase from Aspergillus terreus of interest in winemaking. J Food Sci 66:204–209
Tejirian A, Xu F (2010) Inhibition of cellulase-catalyzed lignocellulosic hydrolysis by iron and oxidative metal ions and complexes. Appl Environ Microbiol 76(23):7673–7682
Michlmayr H, Brandes W, Eder R, Schümann C, del Hierro AM, Kulbe KD (2011) Characterization of two distinct Glycosyl hydrolase family 78 alpha-l-rhamnosidases from Pediococcus acidilactici. Appl Environ Microbiol 77(18):6524–6530
Acknowledgments
The authors gratefully acknowledge financial support of CIUNT, ANPCYT, and CONICET. They are also grateful for the technical assistance provided by Dr. Ricardo Fitzsimons and Dr. Emilio Rodriguez. We would also like to thank Prof. Mirta Daz at the Universidad Nacional de Salta for donating the naringin, prunin, and naringenin used in the present work.
Conflict of interest
None.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is dedicated in memoriam to Prof. Carlos M. Abate, colleague and friend.
Rights and permissions
About this article
Cite this article
Alvarenga, A.E., Romero, C.M. & Castro, G.R. A novel α-l-rhamnosidase with potential applications in citrus juice industry and in winemaking. Eur Food Res Technol 237, 977–985 (2013). https://doi.org/10.1007/s00217-013-2074-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-013-2074-y