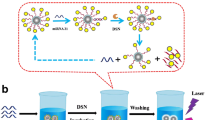
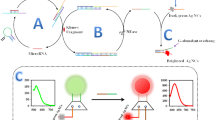
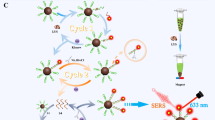
Abstract
Here, we developed a surface-enhanced Raman scattering (SERS) sensor based on functionalized Au@Ag core–shell nanorods (Au@Ag NRs) and cascade DNAzyme amplifier (CSA) for sensitive and accurate determination of microRNA-21 (miRNA-21). The as-prepared SERS nanoprobes were composed of a thiol-modification hairpin probe (HP2)–functionalized Au@Ag NRs and hairpin DNAzyme (HP1-Dz). Compared with original gold nanorods, the silver shell caused an enhancement of plasmonic properties, resulting in a significant enhancement of Raman signals. In the presence of target miRNAs, the hairpin construction of HP1-Dz changed due to DNA/RNA hybridization; subsequently, the DNAzyme-catalyzed cleaving process changed, and the Raman signals of the SERS nanoprobes gradually “turned off” with time elapse because of the dissociation of the Raman reporter from the surface of Au@Ag NRs. Hence, based on this principle, the proposed SERS sensor exhibited good linearity in the range 0.5 fM to 10 nM for miRNA-21 detection with a detection limit (LOD) of 0.5 fM. The proposed SERS platform has potential application in quantitative and precise detection of miRNA-21 in human serum.
Graphical abstract








Similar content being viewed by others
References
Mallory AC, Vaucheret H. Functions of microRNAs and related small RNA in plants. Nat Genet. 2006;38:S31–6.
Chinnappan R, Mohammed R, Yaqinuddin A, Abu-Salah K, Zourob M. Highly sensitive multiplex detection of microRNA by competitive DNA strand displacement fluorescence assay. Talanta. 2019;200:487–93.
Zhou W, Tian YF, Yin BC, Ye BC. Simultaneous surface-enhanced Raman spectroscopy detection of multiplexed microRNA biomarkers. Anal Chem. 2017;89:6120–8.
Zheng J, Ma DD, Shi ML, Bai JH, Li YH, Yang JF, et al. A New Enzyme-free quadratic SERS signal amplification approach for circulating microRNA detection in human serum. Chem Commun. 2015;51:16271–4.
Qiu X, Wang P, Cao ZJ. Hybridization chain reaction modulation DNA-hosted silver nanoclusters for fluorescent identification of single nucleotide polymorphisms in the let-7 miRNA family. Biosens Bioelecron. 2014;60:351–7.
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179.
Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175.
Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat Methods. 2004;1:155–61.
Laing S, Gracie K, Faulds K. Multiplex in vitro detection using SERS. Chem Soc Rev. 2016;45:1901–18.
Etchegoin PG, Lacharmoise PD, Le Ru EC. Influence of photostability on single-molecule surface enhanced Raman scattering enhancement factors. Anal Chem. 2009;81:682–8.
Fan XC, Hao Q, Li MZ, Zhang XY, Yang XZ, Mei YF, et al. Hotspots on the move: active molecular enrichment by hierarchically structured micromotors for ultrasensitive SERS sensing. ACS Appl Mater Inter. 2020;12:28783–91.
Zhou DH, Zeng LW, Pan JF, Li Q, Chen JH. Autocatalytic DNA circuit for Hg2+ detection with high sensitivity and selectivity based on exonuclease III and G-quadruplex DNAzyme. Talanta. 2020;207:120258.
Du SS, Su MK, Jiang YF, Yu FF, Xu Y, Lou XF, et al. Direct discrimination of edible oil type, oxidation, and adulteration by liquid interfacial surface-enhanced Raman spectroscopy. ACS Sensors. 2019;4:1798–805.
Zhou H, Zhang J, Li B, Liu J, Chen HY. Dual-mode SERS and electrochemical detection of miRNA based on popcorn-like gold nanofilms and toehold-mediated strand displacement amplification reaction. Anal Chem. 2021;93(15):6120–7.
Wang SF, Wu CJ, Luo JJ, Luo XX, Yuan R, Yang X. Target-triggered configuration change of DNA tetrahedron for SERS assay of microRNA 122. Microchim Acta. 2020;187:460.
Lee T, Wi JS, Oh A, Na HK, Lee JJ, Lee K, et al. Highly robust, uniform and ultra-sensitive surface-enhanced Raman scattering substrates for microRNA detection fabricated by using silver nanostructures grown in gold nanobowls. Nanoscale. 2018;10:3680–7.
Luo W, Wu CJ, Huang SQ, Luo XL, Yuan R, Yang X. Liquid phase interfacial surface-enhanced Raman scattering platform for ratiometric detection of microRNA 155. Anal Chem. 2020;92:15573–8.
Chen K, Wu L, Jiang XC, Lu ZC, Han HY. Target triggered self-assembly of Au nanoparticles for amplified detection of Bacillus thuringiensis transgenic sequence using SERS. Biosens Bioelecron. 2014;62:196–200.
Xu W, Zhao AW, Zuo FT, Khan R, Hussain HMJ, Li J. A highly sensitive DNAzyme-based SERS biosensor for quantitative detection of lead ion in human serum. Anal Bioanal Chem. 2020;412:4565–74.
Chang J, Zhang AM, Huang ZC, Chen YS, Zhang Q, Cui DX. Monodisperse Au@Ag core-shell nanoparticles with ultrasensitive SERS-activity for rapid identification and Raman imaging of living cancer cells. Talanta. 2019;198:45–54.
Xu W, Zhao AW, Zuo FT, Khan R, Hussain HMJ, Chang JG. Au@Ag core-shell nanoparticles for microRNA-21 determination based on duplex-specific nuclease signal amplification and surface-enhanced Raman scattering. Microchim Acta. 2020;187:384.
Sun YY, Li W, Zhao LQ, Li FY, Xie YF, Yao WR, et al. Simultaneous SERS detection of illegal food additives rhodamine B and basic orange II based on Au nanorod-incorporated melamine foam. Food Chem. 2021;357:129741.
Nguyen L, Dass M, Ober MF, Besteiro LV, Wang ZMM, Nickel B, et al. Chiral assembly of gold-silver core-shell plasmonic nanorods on DNA origami with strong optical activity. ACS Nano. 2020;14:7454–61.
Cui MR, Li XL, Xu JJ, Chen HY. Acid-switchable DNAzyme nanodevice for imaging multiple metal ions in living cells. ACS Appl Mater Inter. 2020;12:13005–12.
Wang R, Xu XW, Li X, Zhang N, Jiang W. pH-responsive ZnO nanoprobe mediated DNAzyme signal amplification strategy for sensitive detection and live cell imaging of multiple microRNAs. Sensor Actuat B-Chem. 2019;293:93–9.
Zhou YJ, Yu SS, Shang JH, Chen YY, Wang Q, Liu XQ, et al. Construction of an exonuclease III-propelled integrated DNAzyme amplifier for highly efficient microRNA detection and intracellular imaging with ultralow background. Anal Chem. 2020;92:15069–78.
He WC, Li ST, Wang LZ, Zhu LJ, Zhang YZ, Luo YB, et al. AuNPs - DNAzyme molecular motor biosensor mediated by neighborhood click chemistry reactions for the ultrasensitive detection of microRNA-155. Sensor Actuat B-Chem. 2019;290:503–11.
Tang LJ, Li S, Han F, Liu LQ, Xu LG, Ma W, et al. SERS-active Au@Ag nanorod dimers for ultrasensitive dopamine detection. Biosens Bioelecron. 2015;71:7–12.
Wu L, Xiao XY, Chen K, Yin WM, Li Q, Wang P. Ultrasensitive SERS detection of Bacillus thuringiensis special gene based on Au@Ag NRs and magnetic beads. Biosens Bioelecron. 2017;92:321–7.
Pang YF, Wang CW, Wang J, Sun ZW, Xiao R, Wang SQ. Fe3O4@Ag magnetic nanoparticles for microRNA capture and duplex-specific nuclease signal amplification based SERS detection in cancer cell. Biosens Bioelecron. 2016;79:574–80.
Xi Q, Zhou DM, Kan YY, Ge J, Wu ZK, Yu RQ, et al. Highly sensitive and selective strategy for microRNA detection based on WS2 nanosheet mediated fluorescence quenching and duplex-specific nuclease signal amplification. Anal Chem. 2014;86:1361–5.
Li YX, Li XH, Meng YC, Hun X. Photoelectrochemical platform for microRNA let-7a detection based on graphdiyne loaded with AuNPs modified electrode coupled with alkaline phosphatase. Biosens Bioelecron. 2019;130:269–75.
Lin XY, Jiang JJ, Wang J, Xia JL, Wang RN, Diao GW. Competitive host-guest recognition initiated by DNAzyme-cleavage cycling for novel ratiometric electrochemical assay of miRNA-21. Sensor Actuat B: Chem. 2021;333:129556.
Zhou Y, Wang HJ, Zhang H, Chai RQ, Yuan R. Programmable modulation of copper nanoclusters electrochemiluminescence via DNA nanocrances for ultrasensitive detection of microRNA. Anal Chem. 2018;90:3543–9.
Zhang H, Fu CP, Yi Y, Zhou XD, Zhou CH, Ying GQ, et al. Magnetic-based SERS approach for highly sensitive and reproducible detection of cancer-related serum microRNAs. Anal Methods. 2018;10:624–33.
Si YM, Xu L, Deng T, Zheng J, Li JS. Catalytic hairpin self-assembly-based SERS sensor array for the simultaneous measurement of multiple cancer-associated miRNAs. ACS Sensors. 2020;5:4009–16.
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 21775064), the Natural Science Foundation of Shandong Province (ZR202103030844), Key Laboratory for Biological Medicine in Shandong Universities, Weifang Key Laboratory for Antibodies Medicine, and the technical support from the Testing Center of Hefei University of Technology.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Written informed consent was obtained, and this study was approved by the Ethics Committee of The First Affiliated Hospital of University of Science and Technology of China (No. 2021-RE-070).
Competing interests
There is no conflict of interest. The authors declare that they have no financial interests that could have appeared to influence the work reported in this paper.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, W., Zhang, Y., Chen, H. et al. DNAzyme signal amplification based on Au@Ag core–shell nanorods for highly sensitive SERS sensing miRNA-21. Anal Bioanal Chem 414, 4079–4088 (2022). https://doi.org/10.1007/s00216-022-04053-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04053-z