Abstract
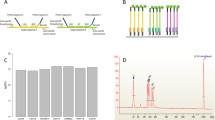
An enzyme-linked oligonucleotide assay (ELONA) for quantification of mRNA expression of five genes involved in breast cancer, extracted from isolated rare tumour cells and amplified by multiplex ligation-dependent probe amplification (MLPA) is presented. In MLPA, a multiplex oligonucleotide ligation assay is combined with a PCR reaction in which all ligation products are amplified by use of a single primer pair. Biotinylated probes complementary to each of the target sequences were immobilised on the surface of a streptavidin-coated microtitre plate and exposed to single-stranded MLPA products. A universal reporting probe sequence modified with horseradish peroxidase (URP–HRP) and complementary to a universal primer used during the MLPA step was further added to the surface-bound duplex as a reporter probe. Simultaneous addition of anchoring probe and target, followed by addition of reporter probe, rather than sequential addition, was achieved with no significant effect on sensitivity and limits of detection, but considerably reduced the required assay time. Detection limits as low as 20 pmol L−1, with an overall assay time of 95 min could be achieved with negligible cross-reactivity between probes and non-specific targets present in the MLPA-PCR product. The same MLPA-PCR product was analysed using capillary electrophoresis, the technique typically used for analysis of MLPA products, and good correlation was observed. The assay presented is easy to carry out, relatively inexpensive, rapid, does not require sophisticated instrumentation, and enables quantitative analysis, making it very promising for the analysis of MLPA products.





Similar content being viewed by others
References
Slade MJ, Payne R, Riethdorf S, Ward B, Zaidi SAA, Stebbing J, Palmieri C, Sinnett HD, Kulinskaya E, Pitfield T, McCormack RT, Pantel K, Coombes RC (2009) Comparison of bone marrow, disseminated tumour cells and blood-circulating tumour cells in breast cancer patients after primary treatment. Br J Cancer 100:160–166
Pantel K, Izbicki JR, Angstwurm M, Braun S, Passlick B, Karg O, Thetter O, Riethmuller G (1993) Immunocytological detection of bone marrow micrometastasis in operable non-small cell lung cancer. Cancer Res 53:1027–1031
Pantel K, Schlimok G, Braun S, Kutter D, Lindemann F, Schaller G, Funke I, Izbicki JR, Riethmuller G (1993) Differential expression of proliferation-associated molecules in individual micrometastatic carcinoma cells. J Natl Cancer Inst 85:1419–1424
Sørensen KM, Andersen PS, Larsen LA, Schwartz M, Schouten JP, Nygren AOH (2008) Multiplex ligation-dependent probe amplification technique for copy number analysis on small amounts of DNA material. Anal Chem 80:9363–9368
Cremonesi L, Ferrari M, Giordano PC, Harteveld CL, Kleanthous M, Papasavva T, Patrinos GP, Traeger-Synodinos J (2007) An overview of current microarray-based human globin gene mutation detection methods. Hemoglobin 31:289–311
Hegde MR, Chin ELH, Mulle JG, Okou DT, Warren ST, Zwick ME (2008) Microarray-based mutation detection in the dystrophin gene. Hum Mutat 29:1091–1099
Cusco I, del Campo M, Vilardell M, Gonzalez E, Gener B, Galan E, Toledo L, Perez-Jurado LA (2008) Array-CGH in patients with Kabuki-like phenotype: identification of two patients with complex rearrangements including 2q37 deletions and no other recurrent aberration. BMC Med Genet 9:27
Konecny M, Zavodna K, Vranova V, Vizvaryova M, Weismanova E, Mlkva I, Kuglik P, Kausitz J, Bartosova Z (2008) Identification of rare complete BRCA1 gene deletion using a combination of SNP haplotype analysis, MLPA and array-CGH techniques. Breast Cancer Res Treat 109:581–583
Huang R, Lin Y, Shi Q, Flowers L, Ramachandran S, Horowitz IR, Parthasarathy S, Huang R-P (2004) Enhanced protein profiling arrays with ELISA-based amplification for high-throughput molecular changes of tumour patients’ plasma. Clin Cancer Res 10:598–609
Anderson KS, Ramachandran N, Wong J, Raphael JV, Hainsworth E, Demirkan G, Cramer D, Aronzon D, Hodi FS, Harris L, Logvinenko T, LaBaer J (2008) Application of protein microarrays for multiplexed detection of antibodies to tumour antigens in breast cancer. J Proteome Res 7:1490–1499
Chu FS, Hsu K-H, Huang X, Barrett R, Allison C (1996) Screening of paralytic shellfish poisoning toxins in naturally occurring samples with three different direct competitive enzyme-linked immunosorbent assays. J Agric Food Chem 44:4043–4047
Farre ML, Oubina A, Marco MP, Ginebreda A, Tirapu L, Barcelo D (1999) Evaluation of 4-nitrophenol ELISA kit for assessing the origin of organic pollution in wastewater treatment works. Environ Sci Technol 33:3898–3904
Sheedy C, Yau KYF, Hirama T, MacKenzie CR, Hall JC (2006) Selection, characterization, and CDR shuffling of naive llama single-domain antibodies selected against auxin and their cross-reactivity with auxinic herbicides from four chemical families. J Agric Food Chem 54:3668–3678
Rolland JM, Apostolou E, de Leon MP, Stockley CS, O’Hehir RE (2008) Specific and sensitive enzyme-linked immunosorbent assays for analysis of residual allergenic food proteins in commercial bottled wine fined with egg white, milk, and nongrape-derived tannins. J Agric Food Chem 56:349–354
Ranki M, Palva A, Virtanen M, Laaksonen M, Soderlund H (1983) Sandwich hybridization as a convenient method for the detection of nucleic-acids in crude samples. Gene 21:77–85
Nickerson DA, Kaiser R, Lappin S, Stewart J, Hood L, Landegren U (1990) Automated DNA diagnostics using an elisa-based oligonucleotide ligation assay. Proc Natl Acad Sci USA 87:8923–8927
Drolet DW, Moon-McDermott L, Romig TS (1996) An enzyme-linked oligonucleotide assay. Nat Biotechnol 14:1021–1025
Fisher M, Harbron S, Taylorson CJ (1997) A system for the quantitation of DNA using a microtiter plate-based hybridization and enzyme amplification technology. Anal Biochem 251:280–287
Ozalp VC, Nygren AOH, O’Sullivan CK (2008) Fluorescent resonance energy transfer (FRET) based detection of a multiplex ligation-dependent probe amplification assay (MLPA) product. Mol Biosyst 4:950–954
Baldrich E, Restrepo A, O’Sullivan CK (2004) Aptasensor development: elucidation of critical parameters for optimal aptamer performance. Anal Chem 76:7053–7063
Baldrich E, Acero JL, Reekmans G, Laureyn W, O’Sullivan CK (2005) Displacement enzyme linked aptamer assay. Anal Chem 77:4774–4784
Campàs M, Katakis I (2006) Characterisation and determination of stability and functionality of biofunctionalised colloidal gold nanoparticles. Anal Chim Acta 556:306–312
Papamichael KI, Kreuzer MP, Guilbault GG (2007) Viability of allergy (IgE) detection using an alternative aptamer receptor and electrochemical means. Sensors and Actuators B: Chemical 121:178–186
Burns DT, Danzer K, Townshend A (2002) Use of the terms “recovery” and “apparent recovery” in analytical procedures - (IUPAC recommendations 2002). Pure Appl Chem 74:2201–2205
Zhou X, Zhou J (2004) Improving the signal sensitivity and photostability of DNA hybridizations on microarrays by using dye-doped core–shell silica nanoparticles. Anal Chem 76:5302–5312
Bowden M, Song LN, Walt DR (2005) Development of a microfluidic platform with an optical imaging microarray capable of attomolar target DNA detection. Anal Chem 77:5583–5588
Marquette CA, Blum LJ (2004) Direct immobilization in poly(dimethylsiloxane) for DNA, protein and enzyme fluidic biochips. Anal Chim Acta 506:127–132
Mallard F, Marchand G, Ginot F, Campagnolo R (2005) Opto-electronic DNA chip: high performance chip reading with an all-electric interface. Biosens Bioelectron 20:1813–1820
Acknowledgements
This work was carried out as part of the Commission of the European communities ICT project Integrated Microsystem for the magnetic isolation and Analysis of Single circulating tumour Cells for Oncology diagnostics and Therapy follow-up, MASCOT, FP6-2004-IST-026752 and partly supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme the (contract FP7-IEF-221198).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Acero Sanchez, J.L., Henry, O.Y.F., Mairal, T. et al. Colorimetric quantification of mRNA expression in rare tumour cells amplified by multiple ligation-dependent probe amplification. Anal Bioanal Chem 397, 2325–2334 (2010). https://doi.org/10.1007/s00216-010-3830-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-010-3830-5