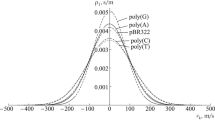
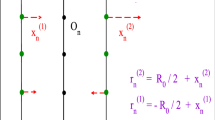
Abstract.
A method is described to extract a complete set of sequence-dependent energy parameters for a rigid base-pair model of DNA from molecular dynamics (MD) simulations. The method is properly consistent with equilibrium statistical mechanics and leads to effective inertia parameters for the base-pair units as well as stacking and stiffness parameters for the base-pair junctions. We give explicit formulas that yield a complete set of base-pair model parameters in terms of equilibrium averages that can be estimated from a time series generated in an MD simulation. The expressions to be averaged depend strongly both on the choice of coordinates used to describe rigid-body orientations and on the choice of strain measures at each junction.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 12 July 2000 / Accepted: 5 January 2001 / Published online: 3 May 2001
Rights and permissions
About this article
Cite this article
Gonzalez, O., Maddocks, J. Extracting parameters for base-pair level models of DNA from molecular dynamics simulations. Theor Chem Acc 106, 76–82 (2001). https://doi.org/10.1007/s002140100256
Issue Date:
DOI: https://doi.org/10.1007/s002140100256