Abstract
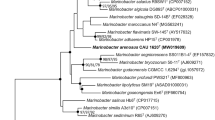
A Gram-negative, aerobic, non-flagellated and ovoid or rod-shaped bacterial strain (JHPTF-M18T), which was isolated from a tidal flat sediment in Republic of Korea, was taxonomically characterized. Strain JHPTF-M18T grew optimally at 25 °C, at pH 7.0–7.5 and in the presence of 2.0–3.0% (w/v) NaCl. 16S rRNA gene sequence analysis showed that strain JHPTF-M18T forms a phylogenetic lineage within the radiation comprising type strains of Mesonia species. The 16S rRNA gene of strain JHPTF-M18T shared sequence similarities of 97.7% with that of type strain of M. mobilis and 92.5–96.8% with those of type strains of the other nine Mesonia species. The DNA G+C content was 33.1% based on its genomic sequence. AAI, ANI and dDDH values between strain JHPTF-M18T and the type strains of M. mobilis, M. hitae, M. oceanica, M. phycicola and M. algae were 72.1–83.7%, 73.1–79.7% and 18.5–22.8%, respectively. Strain JHPTF-M18T contained MK-6 as the predominant menaquinone and iso-C15:0, iso-C17:0 3-OH and summed feature 3 (C16:1 ω7c and/or C16:1 ω6c) as its major fatty acids. Major polar lipids of strain JHPTF-M18T were phosphatidylethanolamine and two unidentified lipids. Strain JHPTF-M18T was separated from recognized Mesonia species by its phenotypic properties together with the phylogenetic and genetic distinctiveness. Based on data presented in this study, strain JHPTF-M18T is considered to represent a novel species of the genus Mesonia. The name Mesonia aestuariivivens sp. nov. is proposed for JHPTF-M18T (=KACC 22185T = NBRC 115119T).

Similar content being viewed by others
Abbreviations
- ANI:
-
Average nucleotide identity
- AAI:
-
Average amino acid identity
- dDDH:
-
Digital DNA–DNA hybridization
- GC:
-
Gas chromatography
References
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kuliko AS, Lesin VM, Nikolenko SI, Phan S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Barrow GI, Feltham RKA (1993) Cowan and steel’s manual for the identification of medical bacteria, 3rd edn. Cambridge University Press, Cambridge
Bernardet J-F (2011) Family I Flavobacteriaceae Reichenbach 1992. In: Krieg NR, Ludwig W, Whitman WB, Hedlund BP, Paster BJ et al (eds) Bergey’s manual of systematic bacteriology, vol 4, 2nd edn. Springer, New York, pp 106–111
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Bruns A, Rohde M, Berthe-Corti L (2001) Muricauda ruestringensis gen. nov., sp. nov., a facultatively anaerobic, appendaged bacterium from German North Sea intertidal sediment. Int J Syst Evol Microbiol 51:1997–2006
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Embley TM, Wait R (1994) Structural lipids of eubacteria. In: Goodfellow M, O’Donnell AG (eds) Modern microbial methods. Chemical methods in prokaryotic systematics. John Wiley & Sons, Chichester, pp 121–161
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Jung YT, Yoon SY, Lee JS, Yoon JH (2016) Taeania maliponensis gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from seawater. Int J Syst Evol Microbiol 66:3552–3557
Kang HS, Lee SD (2010) Mesonia phycicola sp. nov., isolated from seaweed, and emended description of the genus Mesonia. Int J Syst Evol Microbiol 60:591–594
Kim D, Park S, Chun J (2021) Introducing EzAAI: a pipeline for high throughput calculations of prokaryotic average amino acid identity. J Microbiol 59:476–480
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kolberg J, Busse HJ, Wilke T, Schubert P, Kämpfer P, Glaeser SP (2015) Mesonia hippocampi sp. nov., isolated from the brood pouch of a diseased Barbour’s Seahorse (Hippocampus barbouri). Int J Syst Evol Microbiol 2:2241–2247
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102:2567–2572
Lányí B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Lee I, Chalita M, Ha SM, Na SI, Yoon SH, Chun J (2017) ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol 67:2053–2057
Lee SY, Lee MH, Yoon JH (2012) Mesonia ostreae sp. nov., isolated from seawater of an oyster farm, and emended description of the genus Mesonia. Int J Syst Evol Microbiol 62:1804–1808
Leifson E (1963) Determination of carbohydrate metabolism of marine bacteria. J Bacteriol 85:1183–1184
Lucena T, Sanz-Sáez I, Arahal DR, Acinas SG, Sánchez O, Pedrós-Alió C, Aznar R, Pujalte MJ (2020) Mesonia oceanica sp. nov., isolated from oceans during the Tara oceans expedition, with a preference for mesopelagic waters. Int J Syst Evol Microbiol 70:4329–4338
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J (2018) UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
Nedashkovskaya OI, Kim SB, Han SK, Lysenko AM, Rohde M, Zhukova NV, Falsen E, Frolova GM, Mikhailov VV, Bae KS (2003) Mesonia algae gen. nov., sp. nov., a novel marine bacterium of the family Flavobacteriaceae isolated from the green alga Acrosiphonia sonderi (Kütz) Kornm. Int J Syst Evol Microbiol 53:1967–1971
Nedashkovskaya OI, Kim SB, Zhukova NV, Kwak J, Mikhailov VV, Bae KS (2006) Mesonia mobilis sp. Nov., isolated from seawater, and emended description of the genus Mesonia. Int J Syst Evol Microbiol 56:2433–2436
Park S, Won SM, Kim H, Park DS, Yoon JH (2014) Aestuariivita boseongensis gen. nov., sp. nov., isolated from a tidal flat sediment. Int J Syst Evol Microbiol 64:2969–2974
Parte AC (2018) LPSN-List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829
Reichenbach H (1992) The order Cytophagales. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The Prokaryotes, a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd edn. Springer, New York, pp 3631–3675
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J (2015) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 16:681
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Inc, MIDI Technical Note 101. Newark, DE
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Wang FQ, Xie ZH, Zhao JX, Chen GJ, Du ZJ (2015) Mesonia sediminis sp. nov., isolated from a sea cucumber culture pond. Antonie Van Leeuwenhoek 108:1205–1212
Yoon JH, Kim H, Kim IG, Kang KH, Park YH (2003) Erythrobacter flavus sp. nov., a slight halophile from the East Sea in Korea. Int J Syst Evol Microbiol 53:1169–1174
Yoon JH, Lee ST, Kim SB, Kim WY, Goodfellow M, Park YH (1997) Restriction fragment length polymorphism analysis of PCR-amplified 16S ribosomal DNA for rapid identification of Saccharomonospora strains. Int J Syst Bacteriol 47:111–114
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Zhou Y, Gao X, Xu J, Li G, Ma R, Yan P, Dong C, Shao Z (2021) Mesonia hitae sp. Nov., isolated from the seawater of the South Atlantic Ocean. Int J Syst Evol Microbiol 71:4911
Funding
This work was supported by the project on survey of indigenous species of Korea of the National Institute of Biological Resources (NIBR) under the Ministry of Environment (MOE) and “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ015247)” of Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Park, S., Lee, JS., Kim, W. et al. Mesonia aestuariivivens sp. nov., isolated from a tidal flat. Arch Microbiol 204, 550 (2022). https://doi.org/10.1007/s00203-022-03146-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03146-8