Abstract
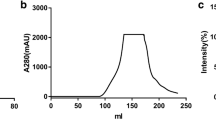
The hemicellulolytic enzyme system produced by Cellulosimicrobium cellulans strain F16 was resolved by ultracentrifugation and size exclusion chromatography. The particle size and molecular weight were determined by both dynamic light scattering and negative stain electron microscopy. The results showed that xylanosomes produced by strain F16 were found to have an apparent sedimentation coefficient of 28 S, were diverse in size (18–70 nm), molecular weight (11–78 MDa) and morphology, but resembled in subunit composition (SDS-PAGE and proteomic results). It is proposed that particles of 22 nm may be the basic unit, while 43 nm and 60 nm particles observed may be dimer and trimer of the basic unit, or xylanosomes with smaller size might be degradation products of larger size xylanosomes. Moreover, such xylanosomes are also found to have strong binding affinity toward water-insoluble substrates such as Avicel, birchwood xylan, and corn cob.





Similar content being viewed by others

Abbreviations
- DLS:
-
Dynamic light scattering
- SEC:
-
Size exclusion chromatography
- DAXP:
-
7-Xylosyl-10-deacetylpaclitaxel
- TEM:
-
Transmission electron microscopy
- MUX:
-
4-Methylumbelliferyl-U-β-d-xylopyranoside
References
Andrews BA, Asenjo JA (1987) Continuous-culture studies of synthesis and regulation of extracellular beta(1–3) glucanase and protease enzymes from Oerskovia xanthineolytica. Biotechnol Bioeng 30:628–637
Artzi L, Bayer EA, Morais S (2017) Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Microbiol 15:83–95
Bajpai P (2014) Chap. 4—structure and synergism between the enzymes of the xylanolytic complex. In: Bajpai P (ed) Xylanolytic enzymes. Academic, Amsterdam, pp 37–41
Bayer EA, Lamed R, White BA, Flint HJ (2008) From cellulosomes to cellulosomics. Chem Rec 8:364–377
Berne BJ, Pecora R (2000) Dynamic light scattering: with applications to chemistry, biology, and physics. Courier Dover Publications, New York
Boonsombuti A, Luengnaruemitchai A, Wongkasemjit S (2013) Enhancement of enzymatic hydrolysis of corncob by microwave-assisted alkali pretreatment and its effect in morphology. Cellulose 20:1957–1966
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Coughlan MP, Honnami K, Honnami H, Ljungdahl LG, Paulin JJ, Rigsby WE (1985) The cellulolytic enzyme complex of Clostridium Thermocellum is very large. Biochem Biophys Res Commun 130:904–909
Doi RH, Kosugi A, Murashima K, Tamaru Y, Han SO (2003) Cellulosomes from mesophilic bacteria. J Bacteriol 185:5907–5914
Dou TY et al (2015a) Functional and structural properties of a novel cellulosome-like multienzyme complex: efficient glycoside hydrolysis of water-insoluble 7-xylosyl-10-deacetylpaclitaxel. Sci Rep 5:13768
Dou TY, Luan HW, Liu XB, Li SY, Du XF, Yang L (2015b) Enzymatic hydrolysis of 7-xylosyltaxanes by an extracellular xylosidase from Cellulosimicrobium cellulans. Biotechnol Lett 37:1905–1910
Erickson HP (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online 11:32–51
Graham J (2001) Biological centrifugation. Springer, Berlin, Heidelberg, New York
Harding S (1999) Protein Hydrodynamics. Protein: a comprehensive treatise, vol 2. JAI Press, Greenwich, pp 271–305
Jiang Z, Dang W, Yan Q, Zhai Q, Li L, Kusakabe I (2006) Subunit composition of a large xylanolytic complex (xylanosome) from Streptomyces olivaceoviridis E-86. J Biotechnol 126:304–312
Mayer F, Coughlan MP, Mori Y, Ljungdahl LG (1987) Macromolecular organization of the cellulolytic enzyme complex of Clostridium Thermocellum as revealed by electron-microscopy. Appl Environ Microbiol 53:2785–2792
Obayashi Y, Iino R, Noji H (2015) A single-molecule digital enzyme assay using alkaline phosphatase with a cumarin-based fluorogenic substrate. Analyst 140:5065–5073
Sunna A, Antranikian G (1997) Xylanolytic enzymes from fungi and bacteria. Crit Rev Biotechnol 17:39–67
Vandooren J, Geurts N, Martens E, Van den Steen PE, Opdenakker G (2013) Zymography methods for visualizing hydrolytic enzymes. Nat Methods 10:211–220
Wang W, Yu Y, Dou TY, Wang JY, Sun C (2018) Species of family Promicromonosporaceae and family Cellulomonadeceae that produce cellulosome-like multiprotein complexes. Biotechnol Lett 40:335–341
Acknowledgements
The authors would like to thank Dr. Hongwei Luan, Xingbao Liu, and Shiyang Li for fermentation and ultrafiltration assistances.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31600641), the National Key Research and Development Program of China (2017YFC1702006), and the Fundamental Research Funds for the Central Universities (No. DUT18RC(4)057).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no direct or indirect conflict of interest.
Additional information
Communicated by Yusuf Akhter.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dou, TY., Chen, J., Hao, YF. et al. Xylanosomes produced by Cellulosimicrobium cellulans F16 were diverse in size, but resembled in subunit composition. Arch Microbiol 201, 163–170 (2019). https://doi.org/10.1007/s00203-018-1606-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-018-1606-z