Abstract
Summary
Osteogenesis imperfecta (OI) is a genetic disease with an estimated prevalence of 1 in 13,500 and 1 in 9700. The classification into subtypes of OI is important for prognosis and management. In this study, we established a clinical severity prediction model depending on multiple features of variants in COL1A1/2 genes.
Introduction
Ninety percent of OI cases are caused by pathogenic variants in the COL1A1/COL1A2 gene. The Sillence classification describes four OI types with variable clinical features ranging from mild symptoms to lethal and progressively deforming symptoms.
Methods
We established a prediction model of the clinical severity of OI based on the random forest model with a training set obtained from the Human Gene Mutation Database, including 790 records of the COL1A1/COL1A2 genes. The features used in the prediction model were respectively based on variant-type features only, and the optimized features.
Results
With the training set, the prediction results showed that the area under the receiver operating characteristic curve (AUC) for predicting lethal to severe OI or mild/moderate OI was 0.767 and 0.902, respectively, when using variant-type features only and optimized features for COL1A1 defects, 0.545 and 0.731, respectively, for COL1A2 defects. For the 17 patients from our hospital, prediction accuracy for the patient with the COL1A1 and COL1A2 defects was 76.5% (95% CI: 50.1–93.2%) and 88.2% (95% CI: 63.6–98.5%), respectively.
Conclusion
We established an OI severity prediction model depending on multiple features of the specific variants in COL1A1/2 genes, with a prediction accuracy of 76–88%. This prediction algorithm is a promising alternative that could prove to be valuable in clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteogenesis imperfecta (OI) is a genetic disease of the connective tissue, and its cardinal feature is bone fragility. Most cases of OI are caused by pathogenic variants in one of the two genes coding for collagen type I alpha chains, either the COL1A1 or the COL1A2 gene [1]. In addition to collagen type I, pathogenic variants in at least 16 other genes have been found to result in OI [2]. The genetic classification has already increased to 20 types of OI (OI types I to XX) (https://omim.org). The prevalence of OI has been estimated at 1 in 13,500 and 1 in 9700 in two recent population-based studies from Scandinavia [3, 4]. Considering all types, OI has a prevalence of approximately 6–7:100,000. COL1A1/2-OI comprises the largest proportion of OI, representing approximately 90% of all cases of OI (www.ncbi.nlm.nih.gov/books/NBK1295) [5].
In clinical practice, the classification into types of OI is important for providing information about prognosis and management, while the primary classification is still based on the clinical manifestations. The Sillence classification describes four OI types (OI type I to IV) [6]; however, the phenotype of OI varies widely between the different types, ranging from mild symptoms with a normal life expectancy to perinatally lethal and progressively deforming symptoms. Nevertheless, the clinical features of different types of OI overlap and subclassification is not an easy task, especially for children in the early stages of the disease without typical clinical symptoms.
Although more than 1000 pathogenic variants have been identified in the COL1A1 and COL1A2 genes, a clear genotype–phenotype correlation has been elusive. Initially, phenotypes resulting from mutations in COL1A1 were thought to be more severe [7]. However, later studies revealed that clinical variations in severity were associated with the affected collagen helical location and types of amino acid substitution [8, 9]. The study of the clinical, molecular, and biochemical consequences of these variants provides unique opportunities to gain insight into the pathophysiological mechanisms underlying OI. We summarized the clinical features of OI patients with different variants in COL1A1 and COL1A2 genes from the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk) and established a prediction model of the clinical severity of OI based on the random forest model. Furthermore, to validate the model, we compared the clinical classification and the results of the prediction model using 17 OI patients from our hospital.
Methods
Data extraction and classification from HGMD as the training set
The HGMD was searched for patients linked to pathogenic/likely pathogenic mutations in the COL1A1 and COL1A2 genes, and the clinical diagnosis with subtype classification and genetic abnormality were collected for each patient. According to the subgroups of genetic abnormalities, these patients were divided into 13 subgroups, including missense variants in glycine (GLY) residue (GLY-ALA, GLY-ARG, GLY-ASP, GLY-CYC, GLY-GLU, GLY-SER, GLY-VAL), missense variants in non-GLY residue (non-GLY), nonsense variants, del < 10 bp, del > 10 bp, ins < 10 bp, and ins > 10 bp subgroups for the subsequent genotype–phenotype correlation analysis. Regarding the severity, we classified OI types II, III, and II/III as the lethal/severe group and OI types I, IV, I/IV, and III/IV as the mild/moderate group.
Prediction features
Initial feature extraction focus on variant type only
According to previous studies, the clinical variations in severity were associated with the types of the variants in COL1A1/COL1A2. Initially, we extracted features according to the type of variants to construct the prediction models, including chromosome position, located codon, amino acid change, and amino acid substitution.
Complement of other features
Apart from the previously reported features that focused on the types of variants, we further included other features to complement the description of the variants and trained the prediction models together with initial extracted features. Specifically, the selected features included the conservation information of constrained coding regions (CCRs) [10] and phastCons conserved elements [11], secondary structure, domain information, motif information, variant type, and gene structure. Details of the complemented features used for the clinical subtype prediction of patients with COL1A1 and COL1A2 pathogenic variants are provided in Supplementary Table 1.
Random forest model for clinical severity prediction
To predict the clinical severity of patients with COL1A1/COL1A2 pathogenic variants, we respectively established prediction models using the selected training set data based on the random forest model, a commonly used machine learning algorithm that combines the output of multiple decision trees to reach a single result and is capable of handling classification [12]. The training set was obtained from the HGMD, and the classification labels, which included lethal/severe OI and mild/moderate OI, were respectively referred to as “1” and “0” in the prediction model. The training of the prediction model was performed with default parameters, using 90% as the training set and the remaining 10% of the data to perform validation in the COL1A1 prediction model, and 80% for training and the remaining 20% for validation in the COL1A2 prediction model.
Evaluation scenarios
Feature importance evaluation
For using variant-type features only and complement other feature prediction models, we both calculated the Gini importance, the total reduction of the criterion brought by the feature, to sort the features in descending order of importance. Specifically, the Gini importance for each predictive feature (f) was measured by the total decrease in the Gini index (improvement in node purity) for all splits on that feature, averaged over all trees in the forest. The measurement was the underlying implementation for the default “feature_importances” method available in the Python scikit-learn library. We used the calculated Gini importance and obtained the optimal subset of features.
with the Gini index (GI) for a specific node (n) as the sum of the variance in proportion for all classes: \(\sum_{k\ne {k}^{*}}{P}_{nk}{P}_{n{k}^{*}}=\sum_{k=1}^{k}{P}_{nk}(1-{P}_{nk})\),
where \({P}_{nk}\) was the proportion of the class k observations in node n.
Prediction performance evaluation
To assess the binary classification performance of our prediction model, the threshold for binary classes, true positive (TP), false negative (FN), true negative (TN), and false positive (FP), was calculated. And the metrics of binary assessment: accuracy (ACC) was computed as ACC = (TP + TN)/(TP + TN + FP + TN). Additionally, the two broadly employed measurements to estimate the prediction performance including the receiver operator characteristics curve (ROC) and the area under the ROC curve (AUC) were also derived from the prediction model. The ROC curve is plotted with the false-positive rate against the true-positive rate. The more the AUC is closer to the maximum value of 1, the better the predictor is, and the value of the AUC of random guessing is usually close to 0.5.
OI patients from our hospital as the test set
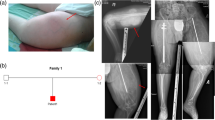
In this study, patients who carried pathogenic or likely pathogenic variants in the COL1A1 or COL1A2 gene between January 1, 2017, and December 31, 2019, and met at least 1 of the following five clinical criteria were included: (1) fracture; (2) skeletal deformity or osteoporosis; (3) family history of OI; (4) short stature, (defined as W and L more than 2 SDS below the normal mean for age and sex [13]); and (5) blue sclerae. The Sillence principle was used for the clinical classification of OI types (OI type I to IV) (6). Patients were excluded if they had pathogenic variants in other genes or pathogenic copy number variants.
Genomic DNA fragments of patients were enriched for exome sequences using the Agilent (Santa Clara, CA, USA) SureSelectXT Human All Exon 50 Mb kit. The DNA libraries were sequenced on a HiSeq2500 sequencer (Illumina, San Diego, CA) according to the manufacturer’s instructions. Variants were annotated by ANNOVAR and VEP software. All variants were classified based on the ACMG guideline [14]. The detected variants were confirmed using PCR, and PCR-amplified DNA products were subjected to direct automated sequencing (3500XL Genetic Analyzer, Applied Biosystems) according to the manufacturer’s specifications.
The samples used in this study were collected with appropriate informed consent and approval of the ethics committee of Children’s Hospital, Fudan University. The methods used in this study were carried out in accordance with the approved guidelines.
Statistical tests
The clinical features of our cases carrying COL1A1/2 variants and the patients with COL1A1/2 pathogenic variations reported in the HGMD were compared. Differences in the clinical features among different mutation type groups were analyzed using Fisher’s exact test. A two-sided P value was used to test for statistical significance (P < 0.05).
Results
Filtering and selection of records in the HGMD
A total of 1338 records were identified from the HGMD, and 252 records without a clinical subtype classification, 60 CNVs involving the non-coding region or multiple exons, and 236 variants in the splice site or non-coding region were removed. Finally, a total of 790 records were selected for further analysis, including 472 records of the COL1A1 gene and 318 records of the COL1A2 gene. The flow chart of records in the HGMD screening and selection process is shown in Fig. 1.
Variant types in different clinical subgroups in the HGMD
COL1A1
According to the clinical subgroup classification, among the 472 patients with a COL1A1 gene defect, 176 (37.3%) patients were classic non-deforming OI with blue sclerae (previously OI type I), 137 (29.0%) patients were perinatally lethal OI (previously OI type II), 64 (13.6%) patients were progressively deforming OI (previously OI type III), 63 (13.3%) patients were common variable OI with normal sclerae (previously OI type IV), 7 (1.5%) patients were I/IV, 5 (1%) patients were II/III, and 20 (4.2%) patients were III/IV (Supplementary Table 2). OI type I was the most common clinical subgroup.
According to the subtypes of genetic abnormalities, among the 472 patients with a COL1A1 abnormality, 270 (57.2%) were the GLY subtype, 41 (8.7%) non-GLY subtype, 26 (5.5%) NS subtype, 127 (26.9%) del/ins < 10 bp subtype, and 8 (1.7%) del/ins > 10 bp subtype (Supplementary Table 2). The missense variants in GLY residue were the most common variant subtype.
According to severity, we classified OI type II, III, and II/III as the lethal/severe group and I, IV, I/IV, and III/IV as the mild/moderate group. The lethal/severe OI group (II, III, and II/III) accounted for 43.6% of all patients with COL1A1 gene pathogenic variants in the HGMD (Fig. 1). The proportion of missense variants in the GLY residue was 80.1% in the lethal/severe OI group and 39.5% in the mild/moderate OI group, and the difference was statistically significant (p < 0.0001). The proportion of nonsense variants was 0.5% in the lethal/severe OI group and 9.4% in the mild/moderate OI group, the proportion of del/ins variants was 8.7% in the lethal/severe OI group and 44.0% in the mild/moderate OI group, and the difference in both was statistically significant (p < 0.0001). For patients with COL1A1 gene defects, missense variants in GLY residue were more likely to cause severe OI, while nonsense and frameshift variants were more likely to cause mild/moderate OI.
COL1A2
Among the 318 patients with a COL1A2 gene defect, 55 (17.3%) patients were OI type I, 61 (19.2%) patients were OI type II, 81 (25.5%) patients were OI type III, 89 (28.0%) patients were OI type IV, 6 (1.9%) patients were I/IV, 9 (2.8%) patients were II/III, and 17 (5.3%) patients were III/IV (Supplementary Table 3). OI type IV was the most common clinical subgroup.
Among the 318 patients with a COL1A2 abnormality, 275 (86.5%) were the GLY subtype, 18 (5.7%) non-GLY subtype, 2 (0.6%) NS subtype, 15 (4.7%) del/ins < 10 bp subtype, and 8 (2.5%) del/ins > 10 bp subtype (Supplementary Table 3). The missense variants in GLY residue were the most common.
The lethal/severe OI group (II, III, and II/III) accounted for 47.5% of all patients with COL1A2 gene pathogenic variants in the HGMD (Fig. 1). The proportion of missense variants in the GLY residue was 88.1% in the lethal/severe OI group and 85.0% in the mild/moderate OI group, the proportion of nonsense variants was 0 in the lethal/severe OI group and 1.2% in the mild/moderate OI group, and the proportion of del/ins variants was 8.6% in the lethal/severe OI group and 6.0% in the mild/moderate OI group. There were no statistical differences in the proportion of any types of variation between lethal/severe OI and mild/moderate OI. For patients with COL1A2 gene defects, missense variants in GLY residue accounted for the largest proportion (86.5%). However, there was no statistical difference between the type of variant and the disease severity.
Prediction results based on the types of variants only
The features of COL1A1 and COL1A2 initial prediction models were extracted according to the subgroups of genetic abnormalities as described in the “Methods” section. Specifically, the initial prediction for COL1A1 and COL1A2 included 13 and 14 features, respectively.
We used the training set data of the COL1A1 and COL1A2 pathogenic variants obtained from the HGMD (19 and 6 pathogenic variants respectively in COL1A1 and COL1A2 that lack conservation information were not included in the prediction) (Supplementary Table 2 and Supplementary Table 3), and the default parameters were used to train and build the prediction model with the supplemented features of the COL1A1 and COL1A2 variants described in the “Methods” section (Figs. 2B and 3B).
Distribution of HGMD samples with COL1A1 pathogenic variants, relative importance of individual features, and the ROC of the COL1A1 prediction model in the training set. A The located codon and the variation type are plotted according to the HGMD records with COL1A1 pathogenic variants. Each square represents a single patient, with colors reflecting the patients’ clinical types. B Relative importance of individual features in the prediction model with variant-type features only and optimized features. Gini importance estimates were normalized to sum to one, and the importance of an individual feature reflects its intrinsic predictive ability. C The AUC of predicting a patient with a pathogenic COL1A1 variant to be lethal to severe OI or mild to moderate OI in the HGMD training set; the AUC of the prediction is 0.767 and 0.902% with variant-type features only and optimized features, respectively
Distribution of HGMD samples with COL1A2 pathogenic variants, relative importance of individual features, and the ROC of the COL1A2 prediction model in the training set. A The located codon and the variation type are plotted according to the HGMD records with COL1A2 pathogenic variants. Each square represents a single patient, with colors reflecting the patients’ clinical types. B Relative importance of individual features in the prediction model with variant-type features only and optimized features. Gini importance estimates were normalized to sum to one, and the importance of an individual feature reflects its intrinsic predictive ability. C The AUC of predicting a patient with a pathogenic COL1A2 variant to be lethal to severe OI or mild to moderate OI in the HGMD training set; the AUC of the prediction is 0.545 and 0.731% with variant-type features only and optimized features, respectively
For the clinical severity prediction of patients with a COL1A1 pathogenic variant, the training set data included 453 samples, with 201 and 252 samples respectively labeled as lethal/severe OI and mild/moderate OI. The prediction results showed that the area under the receiver operating characteristic curve (AUC) for predicting lethal/severe OI or mild/moderate OI was 0.767 (Fig. 2C).
As for the prediction of patients with a COL1A2 pathogenic variant, the training set data altogether included 312 samples, with 165 and 147 samples respectively labeled as lethal/severe OI and mild/moderate OI. The prediction results showed that the AUC for predicting lethal/severe OI or mild/moderate OI was 0.545 (Fig. 3C).
Features optimization
We observed that some variant-describing features showed different distributions between the lethal/severe OI group and the mild/moderate OI group. Therefore, we further added these features for COL1A1 and COL1A2 prediction (Supplementary Table 1). The COL1A1 prediction further combined other 17 individual scores (features), including three conservation scores, eight secondary structure/domain/modification/motif scores, and six mutation type scores. Similarly, COL1A2 prediction involved other 13 individual scores, including two conservation scores, six functional scores corresponding to COL1A1, and additional five mutation type scores (Supplementary Table 2 and Supplementary Table 3). Besides, since several mutations in the obtained HGMD training set lacked the selected conservation features, 19 and 6 patients together with their variants in COL1A1and COL1A2 were respectively excluded in the prediction performances.
We calculated the Gini importance of each feature in the initial and feature add prediction models of both COL1A1 and COL1A2. As shown in Figs. 2B and 3B, the conservation features were the most important features in both COL1A1 and COL1A2 models, demonstrating that the added features contributed a lot to the OI clinical severity prediction compared with traditional consideration of variant type alone. Besides, there were two features respectively in COL1A1 and COL1A2 that showed zero importance, which was because only few data in the obtained HGMD training set had these features (N-terminal region n = 1 and ins > 10 bp n = 1 in the COL1A1 training set; stopgain n = 2 and ins > 10 bp n = 1 in the COL1A2 training set).
Prediction results with feature optimization
The prediction results showed that the AUC for predicting lethal/severe OI or mild/moderate OI with COL1A1 pathogenic variant was 0.902 using optimized features (Fig. 2C), which was better than the 0.767 using variant-type features only.
As for the prediction of patients with a COL1A2 pathogenic variant, the prediction results showed that, with feature optimization, the AUC for predicting lethal/severe OI or mild/moderate OI was 0.731, which was better than 0.545 using variant-type features too (Fig. 3C). Detailed information of the training set features used for the prediction of COL1A1 and COL1A2 pathogenic variants is provided in Supplementary Table 2 and Supplementary Table 3.
The 22 patients from our hospital as the test set
A total of 22 unrelated patients (male: 17, female: 5) met the inclusion criteria and were enrolled; 14 of these patients were identified with pathogenic/likely pathogenic variants in the COL1A1 gene and 8 with variants in the COL1A2 gene. According to the clinical severity-classification criteria, ten patients [1, 3, 8, 11, 15, 17,18,19,20, and 22] were classified as lethal/severe, and the other twelve patients were classified as the mild/moderate group (Table 1).
Among the twenty-one pathogenic/likely pathogenic variants, we detected 14 reported pathogenic variants and 7 novel variants. The 21 variants included 10 missense variants (7 GLY residues, 3 non-GLY residues), 5 frameshifts, 2 nonsense variants, and 4 splice acceptor variants (Table 2). Missense variants accounted for 47.6% of the variants in this study. Among all 7 novel variants, 4 of them were frameshift, 1 splice acceptor variant, and 2 missense variants.
Prediction results of 17 patients from our hospital
Based on the prediction model established in the training set, we also predicted the clinical subgroup of the 17 patients from our hospital, except 4 patients with splicing site variant and 1 patient without complementary conservation features. The prediction result of using variant-type features and complemented features were 13 and 15 patients, respectively, consistent with the clinical manifestation. The prediction results showed an overall accuracy of 76.5% (95% CI: 50.1–93.2%) and 88.2% (95% CI: 63.6–98.5%), respectively.
Of the 10 patients with a COL1A1 mutation, 3 were classified as lethal/severe OI and 7 were classified as mild/moderate OI. The prediction result of using variant-type features and complemented features were 8 and 9 patients, respectively, consistent with the clinical manifestation, with respective prediction accuracy of 80.0% (8/10) and 90% (9/10). One patient [3] with severe phenotypes was predicted to be mild/moderate. Detailed information of the 10 patients is provided in Supplementary Table 4.
Of the 7 patients with a COL1A2 mutation, 5 were classified as lethal/severe OI and 2 were classified as mild/moderate OI. The prediction result of using variant-type features and complemented features were 5 and 6 patients, respectively, consistent with the clinical manifestation, with respective prediction accuracy of 71.4% (5/7) and 85.7% (6/7). One patient [21] with mild phenotypes was predicted to be lethal/severe. Detailed information of the 7 patients is provided in Supplementary Table 5.
Discussion
General rules for genotype–phenotype correlations in COL1A1/2-OI have been published [5]. Mutations in COL1A1 and COL1A2 can result in the haploinsufficiency of type I collagen (quantitative defect) or structural defect (qualitative defect) [5, 8, 15]. Classic non-deforming OI almost results from a pathogenic variant in one COL1A1 and COL1A2 allele that introduces a quantitative defect of the collagen type I alpha 1 chain. Quantitative defects can result from nonsense variants, frameshift mutations, the splice site, and deletions of the entire COL1A1/2 gene [16,17,18]. Structural collagen defects (qualitative defect), often caused by glycine substituting, can cause lethal type II, type III, and type IV OI [8].
In this study, we summarized the variant types in different clinical subgroups in the HGMD, finding that the proportion of missense variants in GLY residue in the COL1A1 gene was higher in the lethal/severe OI group than in the mild/moderate OI group. Glycine is the least bulky amino acid, and other substituting amino acids do not fit well into the collagen triple helix [19]. This is in keeping with the predicted theory that the greater the disruption of the triple helix assembly, the more severe the phenotype presents [20, 21]. Structural studies analyzing the effects of glycine substitutions with bulkier, more hydrophilic amino acid side chains on collagen triple helix assembly have shown progressive disruption of the triple helix and molecular assembly [22, 23]. In this study, however, we found no statistical differences in the proportion of any variation type in the COL1A2 gene between lethal/severe and mild/moderate OI. This prompted us to try to develop a prediction model of clinical severity.
Previous extensive studies provided abundant information for us to build a prediction model of disease severity. Schleit et al. reported a study focusing on the relationships between splicing variants in the COL1A1 gene and patients’ phenotype, finding that the prediction results were only correct in 42–74% of instances by established prediction tools [16]. General predictors like SIFT, Polyphen2, and MutationTaster have made great progress in predicting variant pathogenicities. The strength of these predictors is that they were trained on large datasets, along with powerful learning techniques capturing general mutational pathogenicity information. Although adding gene-specific biological context can improve the prediction effect for the gene-specific prediction model, due to the limited data, there are currently few tools available for specific gene pathogenicity predictions. In this study, we established a predictive method for OI severity caused by pathogenic variants in the COL1A1/2 genes. To the best of our knowledge, this is the first study aimed to establish a prediction model based on the genotype–phenotype correlation from a large public database to predict the severity of this disorder.
We initially established a random forest prediction model depending on features related to the variant’s type only. For the training set, the prediction results showed that the AUC was 0.767 and 0.545 for COL1A1 and COL1A2 defects, respectively (Figs. 2C and 3C). Next, we observed that some variant-describing features showed different distributions between the lethal/severe and the mild/moderate OI groups (Supplementary Table 1). Therefore, we built the prediction model with optimized features. After optimization, the prediction results showed a respective AUC of 0.902 and 0.731 for COL1A1 and COL1A2 defects in the training set, and a respective AUC of 0.90 and 0.857 for COL1A1 and COL1A2 mutations in the test set. This model predicted the clinical severity examined with a high degree of accuracy. At present, the treatment of OI is largely dependent on the clinical severity. For children with lethal/severe OI, their parents and other caregivers should be instructed with more details in safe handling techniques. For children themselves, not only contact sports should be avoided, but physical activity should be strictly limited. In addition, the use of bracing to try to stabilize progressively deforming limbs also depends on the subtype of OI. Accurate prediction of clinical severity is beneficial to precision medicine.
In the clinical application of the prediction model, it should be noted that the accuracy may vary with different types of variants. The clinical severity prediction was accurate for all missense variants in the GLY residue in the COL1A1 gene. One patient [3] with severe phenotype carrying a novel COL1A1 frameshift variant (c.3291_3294dup) was predicted to be in the mild/moderate group. One patient [21] with mild phenotype carrying a COL1A2 novel missense variant (c.2108G > T(p.G703V)) was predicted to be in the lethal/severe group. In the presented prediction model, the location and mutation type are two essential features. The accuracy of the prediction of missense variation is relatively high. For the available training set data, only limited data were frameshift variants (the same as case 3), which might explain the inaccurate prediction of the frameshift variant in this study. Although the overall accuracy is relatively high, it is still challenging for prenatal diagnosis.
A disparity in the predictive accuracy between COL1A1 and COL1A2 variants was observed. Generally, the prediction performance is related to the training set data, selected features, and prediction models. Since both COL1A1 and COL1A2 variants used the random forest model in this study, the performance difference shall be caused by the training set data and the used features. Specifically, for the training set data, the differences are reflected in (1) different enrolled numbers of COL1A1 and COL1A2 variants and (2) different distribution patterns of two genes’ variants (Figs. 2A and 3A) and unbalanced outcome data provided by HGMD (more lethal/severe data). As for the features, although the feature categories of COL1A1 and COL1A2 were generally consistent, detailed used features were slightly different according to the gene/variant characters and available information. The number of detailed used features of COL1A1 and COL1A2 were 17 and 15, respectively, with an intersection of 11 consistent features (Supplementary Table S1). These differences are likely to result in the disparity in prediction accuracy between COL1A1 and COL1A2 variants.
In clinical practice, for diseases with a clinical phenotype spectrum like OI, it is important to predict a continuous range of severity. However, the clinical phenotypes of OI are rather complex and the performance of machine learning is highly dependent on the available data. It is important to note that the criteria of OI clinical severity used in the training set are the Sillence categories. First, there may be phenotypic overlap in the specific categories, and second, OI-IV is a common variable OI subtype. In this study, we chose to binarize the OI outcomes considering the limited training data. As shown in Supplementary Table 1, the available training set was inadequate and unbalanced in number for robust prediction model constructions of all OI subtypes. Considering that the mild/moderate and lethal/ severe classifications were practical for both clinicians and patients, we binarized the outcomes and used the ROC for prediction performance evaluation. It is reasonable to believe that the accumulated qualified training data, optimized features, and robust prediction models could help achieve spectrum outcome predictions. And the confusion matrix could be a good alternative metric. Genotype–phenotype relationships and molecular mechanisms for type I collagen-related inherited diseases will be further enriched and revealed as the number of rare disease cases increases.
For patients with COL1A1 gene defects, missense variants in GLY residue were more likely to cause severe OI. However, there was no statistical difference between the type of variant and the disease severity in patients with a COL1A2 gene defect. Therefore, we built a prediction model with more optimized features, rather than just depending on the type of variants. This prediction algorithm is a promising alternative that could prove to be valuable in clinical practice.
References
Bardai G, Moffatt P, Glorieux FH et al (2016) DNA sequence analysis in 598 individuals with a clinical diagnosis of osteogenesis imperfecta: diagnostic yield and mutation spectrum. Osteoporosis international : a journal established as result of cooperation between the European Foundation for Osteoporosis and the National Osteoporosis Foundation of the USA 27(12):3607–3613
Tauer JT, Robinson ME, Rauch F. Osteogenesis imperfecta: new perspectives from clinical and translational research. JBMR Plus. 2019;3(8):e10174.
Folkestad L. Mortality and morbidity in patients with osteogenesis imperfecta in Denmark. Dan Med J. 2018;65(4).
Lindahl K, Astrom E, Rubin CJ et al (2015) Genetic epidemiology, prevalence, and genotype-phenotype correlations in the Swedish population with osteogenesis imperfecta. Eur J Hum Genet 23(8):1112
Ben Amor IM, Glorieux FH, Rauch F. Genotype-phenotype correlations in autosomal dominant osteogenesis imperfecta. J Osteoporos. 2011;2011:540178.
Sillence DO, Rimoin DL (1978) Classification of osteogenesis imperfect. Lancet 1(8072):1041–1042
Prockop DJ, Constantinou CD, Dombrowski KE et al (1989) Type I procollagen: the gene-protein system that harbors most of the mutations causing osteogenesis imperfecta and probably more common heritable disorders of connective tissue. Am J Med Genet 34(1):60–67
Marini JC, Forlino A, Bachinger HP et al (2017) Osteogenesis imperfecta Nat Rev Dis Primers 3:17052
Symoens S, Hulmes DJ, Bourhis JM et al (2014) Type I procollagen C-propeptide defects: study of genotype-phenotype correlation and predictive role of crystal structure. Hum Mutat 35(11):1330–1341
Havrilla JM, Pedersen BS, Layer RM, et al. A map of constrained coding regions in the human genome. Nature Genetics. 2019;51(1):88-+.
Siepel A, Bejerano G, Pedersen JS et al (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15(8):1034–1050
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Li H, Ji CY, Zong XN, et al. Height and weight standardized growth charts for Chinese children and adolescents aged 0 to 18 years. Zhonghua er ke za zhi = Chinese journal of pediatrics. 2009;47(7):487–92.
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics 17(5):405–424
Maioli M, Gnoli M, Boarini M et al (2019) Genotype-phenotype correlation study in 364 osteogenesis imperfecta Italian patients. Eur J Hum Genet 27(7):1090–1100
Schleit J, Bailey SS, Tran T et al (2015) Molecular outcome, prediction, and clinical consequences of splice variants in COL1A1, which encodes the proalpha1(I) chains of type I procollagen. Hum Mutat 36(7):728–739
Bardai G, Lemyre E, Moffatt P et al (2016) Osteogenesis imperfecta type I caused by COL1A1 deletions. Calcif Tissue Int 98(1):76–84
Willing MC, Deschenes SP, Scott DA et al (1994) Osteogenesis imperfecta type I: molecular heterogeneity for COL1A1 null alleles of type I collagen. Am J Hum Genet 55(4):638–647
Marini JC, Forlino A, Cabral WA et al (2007) Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat 28(3):209–221
Sweeney SM, Orgel JP, Fertala A et al (2008) Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J Biol Chem 283(30):21187–21197
Orgel JP, San Antonio JD, Antipova O (2011) Molecular and structural mapping of collagen fibril interactions. Connect Tissue Res 52(1):2–17
Xu P, Huang J, Cebe P et al (2008) Osteogenesis imperfecta collagen-like peptides: self-assembly and mineralization on surfaces. Biomacromol 9(6):1551–1557
Bodian DL, Madhan B, Brodsky B et al (2008) Predicting the clinical lethality of osteogenesis imperfecta from collagen glycine mutations. Biochemistry 47(19):5424–5432
Acknowledgements
We are grateful to the patients and their families for participating in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81873667).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The samples used in this study were collected with appropriate informed consent and approval of the ethics committee of Children’s Hospital, Fudan University. The methods used in this study were carried out in accordance with the approved guidelines. This study was conducted with the consent of the children’s parents and signed informed consent.
Conflicts of interest
None.
Data sharing statement
All the data involved in this study are provided in Supplementary Table 1–5.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
198_2021_6263_MOESM1_ESM.docx
Supplementary Table 1: Origin features used for the clinical subtype prediction of patients with COL1A1 and COL1A2 pathogenic variants. (DOCX 17 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Yang, L., Liu, B., Dong, X. et al. Clinical severity prediction in children with osteogenesis imperfecta caused by COL1A1/2 defects. Osteoporos Int 33, 1373–1384 (2022). https://doi.org/10.1007/s00198-021-06263-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-021-06263-0