Abstract
Proteins of the peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1 (PGC-1) family of transcriptional coactivators coordinate physiological adaptations in many tissues, usually in response to demands for higher nutrient and energy supply. Of the founding members of the family, PGC-1α (also known as PPARGC1A) is the most highly regulated gene, using multiple promoters and alternative splicing to produce a growing number of coactivator variants. PGC-1α promoters are selectively active in distinct tissues in response to specific stimuli. To date, more than ten novel PGC-1α isoforms have been reported to be expressed from a novel promoter (PGC-1α-b, PGC-1α-c), to undergo alternative splicing (NT-PGC-1α) or both (PGC-1α2, PGC-1α3, PGC-1α4). The resulting proteins display differential regulation and tissue distribution and, most importantly, exert specific biological functions. In this review we discuss the structural and functional characteristics of the novel PGC-1α isoforms, aiming to provide an integrative view of this constantly expanding system of transcriptional coactivators.
Similar content being viewed by others
Introduction
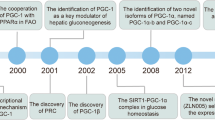
It is now more than 15 years since the cloning of the transcriptional coactivator known as peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) [1]. Since then, PGC-1α and its homologues, PGC-1β and PGC-1-related coactivator (PRC), have been the subject of intense research in a constantly growing number of fields, such as energy metabolism, cardiovascular disease, skeletal muscle physiology, neurodegenerative diseases and even mood disorders. Numerous studies have investigated the roles of PGC-1 coactivators in different physiological and disease settings, and brought to light several mechanisms by which these transcriptional regulators contribute to cellular adaptation to diverse challenges. At the same time, this research has uncovered new levels of complexity in transcriptional regulation mediated by PGC-1 coactivators. Of the three founding members of the family, PGC-1α has received the most attention, partly because of the many and highly regulated mechanisms that determine its activation and biological activity (comprehensively reviewed in [2, 3]). More recently, work from several laboratories has shown that transcription of a single PGC-1α (also known as PPARGC1A) gene is controlled by several promoter regions that, coupled to alternative splicing, give rise to coactivator variants with distinct transcript and protein structure. Alternative promoter usage and alternative splicing are often coupled [4] to increase the complexity of the transcriptome by producing several proteins from an individual gene. This is a universal phenomenon and 86% of the multi-exonic human genes express appreciable levels of at least two populations of distinct mRNAs [5]. The lack of consensus in nomenclature has populated scientific literature with a growing constellation of PGC-1α variants. For clarity, we use PGC-1α/Pgc-1α to refer to the human/rodent gene, PGC-1α when discussing the collective of α proteins, and PGC-1α1 to specify the original protein ([1]; Fig. 1). Here we discuss current advances in our understanding of PGC-1α-related biology, with particular focus on its variants, their structure and biological functions, to give an integrated view of the PGC-1α system of transcriptional coactivators.
PGC-1α isoform structure and transcriptional origin. Transcription initiated from an upstream alternative promoter (AP) of the Pgc-1α gene results in the inclusion of new exons 1b or 1b′ (e1b, e1b′). These encode two distinct N-termini (aa sequence in blue and orange) that are different from the canonical exon 1a (e1a; aa sequence in brown), which follows the proximal promoter (PP). An additional promoter located in intron 2 (LP) regulates expression of a variant in human liver, which includes the novel exon 1L (e1L). A set of brain-specific PGC-1α isoforms are expressed under the control of a promoter region (BP) located 587 kb upstream of e1a. C-terminal amino acid sequences resulting from three different splicing options are shown. *Highly conserved in mammals; transcripts only detected in human tissues
PGC-1s: meet the parents
PGC-1α was first identified in a yeast two-hybrid screen for factors that interact with the nuclear receptor PPARγ in brown adipose tissue (BAT) in the context of cold-induced thermogenesis [1]. Soon after, two homologous genes, PGC-1β [6] and PRC [7] were cloned by cDNA and protein sequence homology, respectively. Although these proteins are encoded by different genes, their modular structure is remarkably similar [8], which explains the partial overlap in their physiological roles [3]. The three PGC-1 homologues share an amino-terminal activation domain with LXXLL signature motifs involved in nuclear receptor docking [6] and interaction with other transcriptional coactivators such as the steroid receptor coactivator-1 (SRC-1)/p300 complex [9], a nuclear localisation signal (NLS), and a C-terminal RNA recognition motif (RRM) [1]. PGC-1α and PRC share an additional carboxy-terminal sequence, the arginine–serine-rich (RS) domain [1], that is characteristic of proteins involved in mRNA processing and splicing [10]. The C-terminal region of PGC-1α1 has been shown to be essential for interaction with the Mediator complex, and thus the transcriptional machinery, via Mediator of RNA polymerase II transcription subunit 1 (MED1) [9]. Additionally, PGC-1α mutants lacking the RS and RRM domains cannot associate with splicing factors and fail to modulate specific splicing events in an in vitro system. This suggests that PGC-1s play a role not only in quantitative regulation of gene expression, but also collaborate with the co-transcriptional processing of nascent mRNAs [11].
Most gene programs regulated by PGC-1α are bioenergetically demanding and result in coordinated cellular actions to adapt to higher energy demands. For this reason, cells keep tight control over PGC-1α activity through diverse transcriptional and post-translational mechanisms. Some of the signalling pathways controlling PGC-1α gene expression involve calcium signalling [12], calcineurin A [13], cyclic AMP [14] and 5′ AMP-activated protein kinase (AMPK) [15]. Activation of these pathways results in the mobilisation of different transcription factors to PGC-1α gene regulatory regions, in a context- and tissue-specific manner. These include several members of the nuclear hormone receptor family, as well as other tissue-specific transcription factors [3]. Access of transcriptional regulators to the appropriate genomic regions can be limited through epigenetic modifications such as DNA methylation [16]. Accordingly, PGC-1α promoter methylation is reduced by physical exercise, a classical inducer of PGC-1α expression in skeletal muscle [17]. Conversely, methylation of the same regions has been found to increase in pathological situations associated with reduced PGC-1α gene expression [18, 19]. In addition, PGC-1α activity can be modulated by a plethora of post-translational modifications, including phosphorylation, acetylation and ubiquitylation. Kinases that target PGC-1α1 include p38 mitogen-activated protein kinase [20], protein kinase B [21], S6 kinase [22], AMPK [23] and glycogen synthase kinase 3β (GSK3β) [24]. These different phosphorylation events constitute a code that changes PGC-1α1 biological activity by, for example, dictating association with specific transcription factors [3]. The acetylation state of PGC-1α1 can be modified by acetyltransferases such as GCN5 [25] or deacetylases like sirtuin 1 (SIRT1) [26, 27]. Additionally, ubiquitylation by specific E3 ligases directly regulates PGC-1α protein stability [24, 28, 29]. Together, these multiple levels of regulation ensure that under any given condition the cellular PGC-1α activity is appropriate to regulate the gene programs necessary to adapt to the new biological environment.
Where do we start? Multiple origins for PGC-1α transcripts
Most of the studies on PGC-1α protein structure and function have focused on the 797 amino acid (aa)-long murine full-length protein (94.7% sequence identity with the 798 aa-long human PGC-1α). However, early on in the PGC-1α history there were reports of other transcripts induced by challenging energy situations, such as cold exposure in BAT [30] and exercise in skeletal muscle [31]. Only recently have various laboratories made efforts to further characterise these PGC-1α variants. By analysing expressed sequence tag (EST) libraries, two independent studies identified two novel Pgc-1α transcripts highly induced in mouse skeletal muscle by exercise and β-adrenergic stimulation [32, 33]. Genomic mapping of the newly identified 5′ sequences revealed that the novel transcripts are expressed from an evolutionarily conserved alternative promoter of the Pgc-1α gene, located approximately 14 kb upstream of the previously characterised transcription start site (TSS) [33]. Transcripts originating from this distal promoter contain a novel exon 1 sequence (exon 1b) that can follow one of two splicing options and thus encode two different amino termini (12 and 3 aa long, respectively; Fig. 1) slightly shorter than the proximal exon 1a (16 aa long; Fig. 1) [32, 33]. Miura et al cloned Pgc-1α-b and Pgc-1α-c (for the long and short exon 1b, respectively) from a muscle cDNA library, thus renaming the original protein as PGC-1α-a [32] (Fig. 1).
PGC-1α-b and PGC-1α-c differ only in their N-termini while the rest of the protein is identical to the canonical PGC-1α1. Concurrently, two other transcripts were identified that contained the long and short exon 1b, those for Pgc-1α2 and Pgc-1α3, but their primary structure was not investigated in this study [33]. Consistent with their nomenclature, the canonical isoform was renamed Pgc-1α1. The alternative gene promoter is highly conserved between species and has been shown to be active in human skeletal muscle [34–36]. Accordingly, the novel PGC-1α isoforms are detected in skeletal muscle after exercise [35–37].
These first studies showed that the proximal Pgc-1α gene promoter, from where Pgc-1α1 is expressed, seems to have higher basal expression whereas the alternative promoter is more responsive to stimulation. Indeed, physical exercise or cold exposure induces a promoter activity shift towards the upstream TSS in skeletal muscle and BAT (respectively), favouring the expression of the novel PGC-1α isoforms [32, 33]. This alternative Pgc-1α gene promoter is highly responsive to β-adrenergic stimulation, both in BAT and skeletal muscle. In line with this, mice treated with clenbuterol (a β2-adrenergic agonist) show levels of expression of the novel PGC-1α isoforms comparable to those induced by exercise training. Conversely, the β2-adrenergic antagonist ICI-118,551 blunts Pgc-1α-b and Pgc-1α-c induction by physical exercise [32], whereas the non-selective β-blocker propranolol significantly reduces the magnitude of muscle Pgc-1α induction by voluntary running [33].
PGC-1α-b and PGC-1α-c are not significantly divergent from PGC-1α1 (i.e. PGC-1α-a) in terms of structure or function. For example, like PGC-1α1 they coactivate PPAR nuclear receptors [32] and all these proteins regulate transcriptional programs that elicit some of the effects of endurance exercise training in skeletal muscle by promoting mitochondrial biogenesis, fatty acid oxidation [32] and angiogenesis [38] (Fig. 2). Chinsomboon and collaborators observed that exercise-mediated induction of Pgc-1α2 and Pgc-1α3 is associated with vascular endothelial growth factor (VEGF) expression and promotion of angiogenesis in an oestrogen-related receptor (ERR)α-dependent manner [33]. Further characterisation of a Pgc-1α-b skeletal muscle-specific transgenic mouse model revealed that the improved oxidative capacity and exercise performance were directly correlated to mitochondrial volume rather than increased mitochondrial function [38]. Pgc-1α-b has also been detected in rat cardiomyocytes after fasting, an event that is dependent on cAMP signalling [39].
PGC-1α isoforms: ABC, L/B, NT and 2, 3, 4
L(iver)- and B(rain)-PGC-1α
In addition to the novel upstream alternative PGC-1α gene promoter [32, 33], other TSSs have been described for the human PGC-1α gene [40, 41]. One site is located within intron 2 of the PGC-1α gene and, despite being in a highly conserved region, it seems to be used only in human fasted liver [40]. The resulting transcript, L-PGC-1α, encodes the novel exon 1L that is spliced to exon 3 and then follows the canonical PGC-1α transcript sequence (Fig. 1). Because the start codon for L-PGC-1α is now located in exon 3, this protein lacks the first 127 aa of PGC-1α1 (encoded by exons 1, 2 and part of 3). Consequently, the first LXXLL motif of the activation domain that facilitates recruitment of SRC-1/p300, and the GCN5 interaction region (aa 1–97 of PGC-1α1) are missing from L-PGC-1α. Since L-PGC-1α maintains the C-terminal NLS, the protein localises in the nucleus, where it coactivates PPARα, PPARγ and hepatocyte nuclear factor 4α (HNF4α) but not liver X receptor α (LXRα) [40]. Like PGC-1α1, L-PGC-1α is induced in response to glucocorticoids and cAMP. It is interesting to note that the lack of aa 1–127 could impact both post-translational modifications, e.g. acetylation, as well as interactions with other transcription factors.
Subsequent work from the same group identified another TSS, located 587 kb upstream of PGC-1α exon 2, which can produce several brain-specific isoforms [41]. In human brain, these novel transcripts seem to be more abundant than the canonical PGC-1α1 and encode several isoforms of varying length that have been confirmed at the transcript level in human brain samples and at the protein level in a SH-SY5Y neuroblastoma cell line [41]. These transcripts contain novel exons B1-5 located upstream of the canonical exon 2. Most of these transcripts include exons B1 followed by B4 and/or B5, and then exons 2–13 [41]. Whether the brain-specific isoforms have functional activation domains remains to be determined. Interestingly, the newly identified promoter that mediates the transcription of these brain PGC-1α isoforms is located in a genomic region associated with age of onset of Huntington’s disease [41], a disease that has been linked to altered PGC-1α function [42–44].
NT(erminal)-PGC-1α
Using a BAT cDNA library, Zhang and collaborators cloned a Pgc-1α variant expressed under the control of the proximal gene promoter that contains an alternative splicing event between exons 6 and 7 [45], which introduces a premature stop codon (Fig. 1). This particular intronic sequence of the Pgc-1α gene, which is highly conserved in mammals, contains two distinct splicing acceptor sites. When the upstream acceptor site is favoured, it yields a 270 aa protein named NT-PGC-1α that corresponds to the activation domain of PGC-1α1 (aa 11–80) and part of the repression domain (aa 180–403 in PGC-1α1; 180–267 in NT-PGC-1α). NT-PGC-1α lacks all the central and C-terminal PGC-1α1 protein modules, including the RS/RRM domains and the NLS [1]. The absence of these sequences unmasks a nuclear export signal (NES) that under basal conditions localises NT-PGC-1α mainly to the cytosol (90%) [45]. Cold exposure or β-adrenergic stimulation of BAT triggers a double mechanism that regulates NT-PGC-1α transcriptional activity. On the one hand, in vitro stimulation of brown adipocytes with a cAMP analogue induces rapid shuttling of cytoplasmic NT-PGC-1α into the nucleus [45]. Additionally, cold exposure induces BAT expression of equivalent levels of full-length and NT-PGC-1α isoforms. The same group investigated which Pgc-1α gene promoter regulates the expression of these inducible isoforms. Two studies showed that stimulation of BAT [46] or skeletal muscle [47] shifts promoter activation, favouring the expression of either full-length or NT-PGC-1α from the novel upstream gene promoter [33]. In this way, the repertoire of Pgc-1α isoforms is increased by the addition of two NT-Pgc-1α variants that contain both versions of the novel exon 1b, namely NT-Pgc-1α-b (which is identical to Pgc-1α4, described by Ruas et al in 2012 [36]) and NT-Pgc-1α-c. Consequently, NT-PGC-1α was renamed NT-PGC-1α-a.
Although NT-PGC-1α proteins lack the C-terminal domain that in PGC-1α1 is required for interaction with the Mediator complex [9], they still coactivate PPARα and PPARγ transcriptional activity [45]. However, this association is strictly ligand-dependent, probably because NT-PGC-1α lacks the amino acid sequences that in PGC-1α1 mediate ligand-independent coactivation of PPARs (aa 338–403 in PGC-1α1) [48]. The lack of C-terminal domains make NT-PGC-1α proteins resistant to Twist-1-mediated inhibition in brown adipocytes [49]. This suggests that, unlike PGC-1α1, NT-PGC-1α cannot be targeted by the negative feedback loop mediated by Twist-1 [50].
The biological activity of NT-PGC-1α-a overlaps considerably with that of PGC-1α1. Indeed, expression of NT-PGC-1α-a or PGC-1α1 in differentiated brown adipocytes drives similar gene programs, with enhanced mitochondrial biogenesis (Fig. 2) and increased carnitine palmitoyltransferase-1b (CPT-1b) and uncoupling protein 1 (UCP1) expression [45]. NT-Pgc-1α-a expression is induced by exercise training, does not depend on β-adrenergic stimulation, and promotes a classical oxidative phenotype in C2C12 cells [47]. Accordingly, the expression profile of NT-Pgc-1α-a is closer to that of Pgc-1α1 and differs from the isoforms transcribed from the alternative gene promoter. NT-Pgc-1α can be found in several tissues and is particularly abundant in mouse brain [45]. NT-Pgc-1α-a and Pgc-1α1 (together with L-PGC-1α in humans) are the only isoforms to date that have been reported to be induced in fasted liver (Fig. 2).
PGC-1α2, -α3 and -α4
Activation of the alternative Pgc-1α promoter located 14 kb upstream of the proximal TSS (Fig. 1) results in the transcription of three additional PGC-1α variants with unique structural features [36]. Two of these variants, Pgc-1α2 and Pgc-1α3, share with Pgc-1α-b and Pgc-1α-c (respectively) the splice variants of exon 1b, but differ vastly in structure from any of the other isoforms (Fig. 1). This is due to a series of splicing events common to both Pgc-1α2 and Pgc-1α3, which eliminate exons 4–6 and 9–13. The splicing of exon 8 to the 3′ untranslated region (UTR) of the Pgc-1α gene then creates a new stop codon common to both transcripts. Consequently, the two resulting proteins (379 and 370 aa long, respectively) retain part of the activation and repression domains and completely lack all the C-terminal motifs of PGC-1α1. The third Pgc-1α isoform described in the same study [36] is 266 aa long and is named PGC-1α4 (Fig. 1). Pgc-1α2, Pgc-1α-b and Pgc-1α4 share the same transcriptional origin (and hence the same N-terminal aa sequence) but Pgc-1α4 undergoes the downstream alternative splicing options seen in the NT-Pgc-1α isoforms (i.e. the inclusion of the new exon 6 and the premature stop codon at 266 aa). Pgc-1α2, -α3 and -α4 mRNA and protein levels are induced by hypertrophic stimuli in skeletal muscle [36, 37] and by cold exposure in BAT, but do not drive the well-known oxidative phenotype induced by other variants [36]. Notably, PGC-1α4 has been shown to exert physiological effects previously unreported for a member of the PGC-1 family. When expressed in skeletal muscle, PGC-1α4 promotes robust muscle hypertrophy by inducing IGF-1 expression and reducing the levels of myostatin, a negative regulator of muscle growth (Fig. 2). Through this mechanism, differential activation of one single Pgc-1α gene in skeletal muscle can result in some of the most-well established benefits of endurance and resistance exercise training (i.e. fatigue resistance and increased muscle mass and strength, respectively). Moreover, high PGC-1α4 levels in mouse skeletal muscle confers resistance to cancer cachexia and disuse-induced atrophy. Muscle hypertrophy mediated by the PGC-1α system is independent of ERRα/γ [36] since PGC-1α4 expression in ERRα or -γ genetically defective myotubes still promotes a hypertrophic phenotype. On the other hand, hypoxia promotes skeletal muscle PGC-1α4 and NT-PGC-1α expression that, together with ERRα, increase VEGF gene transcription and angiogenesis [51]. The biological roles PGC-1α2 and PGC-1α3 remain unknown.
Changes in the PGC-1α system of coactivators in skeletal muscle can crosstalk to other tissues and have systemic effects. Several recent reports demonstrate that elevating specific PGC-1α isoforms in skeletal muscle can impact other tissues by secreting circulating factors (myokines). Irisin and β-aminoisobutyric acid (BAIBA) [52, 53], both under the control of PGC-1α1, or PGC-1α4-regulated myostatin and meteorin-like protein [54, 55], all convey skeletal muscle signals to adipose tissue. Thus, skeletal muscle overexpression of either PGC-1α1 or PGC-1α4 results in browning of adipose tissue, although this is achieved through distinct pathways and mechanisms specific to each isoform.
We have recently uncovered a mechanism by which skeletal muscle PGC-1α1 activation can protect against stress-induced depression. This crosstalk between muscle and the brain is not mediated by secretion of myokines, but results from skeletal muscle detoxification of the tryptophan metabolite kynurenine that upon accumulation in the brain has been linked to neuroinflammation and depression [56].
Challenges and perspectives
Which promoter to promote?
One of the pressing questions about PGC-1α isoform expression is how do cells specifically regulate the different Pgc-1α gene promoters? How do specific stimuli dictate which promoter will be activated? Upon cold exposure, BAT clearly shifts the preference from the proximal to the alternative Pgc-1α promoter and every induced isoform, either full-length or spliced, contains the novel exon 1b [32, 45, 46]. The biological meaning of this specific regulation remains elusive, since the expression of the diverse PGC-1α isoforms in BAT does not seem to trigger a differentiated biological function, and independently of promoter origin or splicing choice, all seem to conduct the mitochondrial thermogenesis program.
Promoter choice and PGC-1α isoform expression is also regulated in skeletal muscle. The 14 kb upstream alternative Pgc-1α gene promoter is highly sensitive to stimulation, and most exercise interventions, from high- to low-intensity endurance training [57] to resistance training [36] seem to activate it to some extent. The classical proximal promoter seems to be less inducible but consistently responsive to high-intensity endurance exercise [57] or freewheel running [33, 58]. Pre-treatment of exercised animals with a β-adrenergic antagonist impairs Pgc-1α-b and Pgc-1α-c induction but does not affect the induction of Pgc-1α1 [32]. In effect, in vivo administration of the β2-adrenergic agonist clenbuterol induces the expression of all isoforms derived from the novel promoter while repressing Pgc-1α1 expression (V. Martínez-Redondo, J. L. Ruas, unpublished observation). This shift adds PGC-1α4-mediated muscle hypertrophy to the bioenergetic effects induced by other alternative isoforms. Understanding how these mechanisms are regulated will allow each of the specific pathways to be targeted therapeutically.
Technical considerations for the detection of PGC-1α isoforms
Since the Pgc-1α gene can generate a variety of mRNAs under different biological conditions and in different tissues, it is important to accurately design the detection reagents when studying specific Pgc-1α isoforms. When performing gene expression analysis by real-time PCR (the most widely used method), it is possible to use isoform-specific combinations of primers that target unique sequences in the different transcripts (Table 1). Moreover, detecting total Pgc-1α levels (which can be done by targeting exon 2, shared by all known isoforms except L-PGC-1α) will, in many cases, mask important changes in the levels of specific isoforms. For example, targeting exon 1a to analyse the effects of clenbuterol administration on mouse muscle PGC-1α expression will detect the overall change in isoforms expressed from the canonical promoter, but fail to measure the induction of the alternative promoter [59]. However, since most transcripts share part of their sequence, real-time PCR is not always a viable approach for reliably measuring PGC-1α isoform expression levels. In many cases, the primer pair combinations that would be truly isoform-specific generate amplicons too large for this technique (i.e. generate non-linear signals). One way of refining PGC-1α isoform analysis is by combining information from two separate primer sets targeting different parts of the transcript and by determining protein molecular sizes by immunoblotting (Table 1). Finally, a third layer of complexity comes from the fact that different PGC-1α isoforms seem to have distinct mRNA or protein stability. For example, equivalent mRNA expression of Pgc-1α1, Pgc-1α2, Pgc-1α3 and Pgc-1α4 in primary myotubes results in different protein levels [36]. Regardless of the transcript proportions in the different Pgc-1α mRNA populations, many of the novel isoforms are more easily detected by immunoblotting than the predominant transcript isoform, PGC-1α1.
Despite the high degree of sequence conservation observed throughout the Pgc-1α gene region, not all the isoforms are expressed in all species. This is the case for the L-PGC-1α isoform, which has only been detected in human tissues [40, 41]. To our knowledge, no available database contains comprehensive information about all the described PGC-1α isoforms. For the human PGC-1α isoforms, Ensembl (www.ensembl.org) (ENSG00000109819) contains entries for PGC-1α1 (001), NT-7a (homologous to mouse NT-Pgc-1α-a) (007), brain-specific isoform 8a (008) and L-PGC-1α (202), which link to the Uniprot (www.uniprot.org) protein database (entry Q9UBK2) as isoforms 1, NT-7a, 8a and 9, respectively. In the same entry are also documented other human brain isoforms B5, B4, B4-8a, B5-NT and B4-3ext (as isoforms 3 to 7). For the mouse variants, only isoforms Pgc-1α1 (001), NT-Pgc-1α-a (002) and Pgc-1α4 (003) can be found in Ensembl (ENSMUSG00000029167), while in Uniprot (entry O70343), isoforms PGC-1α1, -α2, -α3 and -α4 are all documented. The protein sequence of NT-PGC-1α-a can be found in entry Q3LIG2.
As the number of variants described for PGC-1α grows, it would be of great value to agree on a unified, systematic and non-descriptive nomenclature.
A personalised workout for PGC-1s
Another subject of debate is whether different types of exercise training specifically regulate Pgc-1α promoter activation and isoform expression. Alternative promoter-derived Pgc-1α4, which drives muscle hypertrophy, is induced only in exercise programmes that include a resistance component, whereas its expression is unaffected by 8 weeks of aerobic cycling [36]. In addition, a 12-week progressive weight lifting programme specifically induces isoforms Pgc-1α2, Pgc-1α3 and Pgc-1α4 from the novel promoter [37]. However, other research shows that both promoters can be activated independent of the exercise intervention [60, 61]. PGC-1α isoform expression patterns display distinct temporal dynamics [55] and until we understand the mechanisms behind these different expression profiles it is important to unify experimental procedures such as exercise intervention, sampling and gene expression analysis techniques. One common outcome of the exercise training studies with human volunteers is that concurrent training programmes, with both endurance and resistance components, have a synergistic effect on the expression of all PGC-1α isoforms, although the effect is more pronounced on the alternative promoter.
Resistance exercise training protocols are difficult to translate to rodents, so different experimental protocols are used to induce skeletal muscle hypertrophy. Using the hindlimb suspension–reloading model [62] we have seen specific induction of Pgc-1α4 and concomitant repression of Pgc-1α1 during the reloading/hypertrophic phase [36]. In the same study, we demonstrated that PGC-1α4 is sufficient to induce skeletal muscle hypertrophy. A different group has reported that mice lacking expression of all (known) PGC-1α proteins in skeletal muscle still undergo hypertrophy induced by the synergistic ablation model [63]. We cannot rule out either the existence of additional isoforms that might account for those particular effects or compensatory mechanisms due to the disruption of the Pgc-1α gene since the embryonic state. Until we have the appropriate mouse genetic models of PGC-1α isoform-specific gain- and loss-of-function, it will be difficult to establish the in vivo contribution of each protein to the overall effects of PGC-1α activation.
In conclusion, future studies will need to address the mechanisms of regulation and interplay between PGC-1α variants in different tissues and physiological contexts. Finally, we would like to highlight the importance of a more systematic nomenclature for the constantly expanding PGC-1α system of transcriptional coactivators.
Abbreviations
- Aa:
-
Amino acid
- AMPK:
-
5′ AMP-activated protein kinase
- BAT:
-
Brown adipose tissue
- ERR:
-
Oestrogen-related receptor
- NLS:
-
Nuclear localisation signal
- PGC-1α:
-
PPARγ coactivator 1α
- PPARγ:
-
Peroxisome proliferator-activated receptor γ
- PRC:
-
PGC-1-related coactivator
- RRM:
-
RNA recognition motif
- RS:
-
Arginine–serine-rich
- SIRT:
-
Sirtuin
- TSS:
-
Transcription start site
References
Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829–839
Fernandez-Marcos PJ, Auwerx J (2011) Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 93:884S–890S
Villena JA (2015) New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J 282:647–672
Xin D, Hu L, Kong X (2008) Alternative promoters influence alternative splicing at the genomic level. PLoS One 3:e2377
Wang ET, Sandberg R, Luo S et al (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476
Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM (2002) Peroxisome proliferator-activated receptor gamma coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem 277:1645–1648
Andersson U, Scarpulla RC (2001) Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol 21:3738–3749
Lin J, Handschin C, Spiegelman BM (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1:361–370
Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG (2003) Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol Cell 12:1137–1149
Li H, Bingham PM (1991) Arginine/serine-rich domains of the su(w a) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell 67:335–342
Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM (2000) Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell 6:307–316
Wu H, Kanatous SB, Thurmond FA et al (2002) Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296:349–352
Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM (2003) An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc Natl Acad Sci 100:7111–7116
Yoon JC, Puigserver P, Chen G et al (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138
Suwa M, Nakano H, Kumagai S (2003) Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. J Appl Physiol 95:960–968
Cedar H, Bergman Y (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10:295–304
Barres R, Yan J, Egan B et al (2012) Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 15:405–411
Ling C, del Guerra S, Lupi R et al (2008) Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 51:615–622
Barres R, Kirchner H, Rasmussen M et al (2013) Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep 3:1020–1027
Fan M, Rhee J, St-Pierre J et al (2004) Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: modulation by p38 MAPK. Genes Dev 18:278–289
Li X, Monks B, Ge Q, Birnbaum MJ (2007) Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature 447:1012–1016
Lustig Y, Ruas JL, Estall JL et al (2011) Separation of the gluconeogenic and mitochondrial functions of PGC-1α through S6 kinase. Genes Dev 25:1232–1244
Jager S, Handschin C, St-Pierre J, Spiegelman BM (2007) AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A 104:12017–12022
Olson BL, Hock MB, Ekholm-Reed S et al (2008) SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev 22:252–264
Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P (2006) GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3:429–438
Gerhart-Hines Z, Rodgers JT, Bare O et al (2007) Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26:1913–1923
Canto C, Gerhart-Hines Z, Feige JN et al (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458:1056–1060
Wei P, Pan D, Mao C, Wang YX (2012) RNF34 is a cold-regulated E3 ubiquitin ligase for PGC-1α and modulates brown fat cell metabolism. Mol Cell Biol 32:266–275
Trausch-Azar JS, Abed M, Orian A, Schwartz AL (2015) Isoform-specific SCFFbw7 ubiquitination mediates differential regulation of PGC-1α. J Cell Physiol 230:842–852
Kakuma T, Wang ZW, Pan W, Unger RH, Zhou YT (2000) Role of leptin in peroxisome proliferator-activated receptor gamma coactivator-1 expression. Endocrinology 141:4576–4582
Baar K, Wende AR, Jones TE et al (2002) Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J 16:1879–1886
Miura S, Kai Y, Kamei Y, Ezaki O (2008) Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) mRNA in response to β2-adrenergic receptor activation and exercise. Endocrinology 149:4527–4533
Chinsomboon J, Ruas J, Gupta RK et al (2009) The transcriptional coactivator PGC-1α mediates exercise-induced angiogenesis in skeletal muscle. Proc Natl Acad Sci 106:21401–21406
Yoshioka T, Inagaki K, Noguchi T et al (2009) Identification and characterization of an alternative promoter of the human PGC-1α gene. Biochem Biophys Res Commun 381:537–543
Norrbom J, Sällstedt EK, Fischer H, Sundberg CJ, Rundqvist H, Gustafsson T (2011) Alternative splice variant PGC-1α-b is strongly induced by exercise in human skeletal muscle. Am J Physiol Endocrinol Metab 301:E1092–E1098
Ruas JL, White JP, Rao RR et al (2012) A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151:1319–1331
Nader GA, von Walden F, Liu C et al (2014) Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol (1985) 116:693–702
Tadaishi M, Miura S, Kai Y, Kano Y, Oishi Y, Ezaki O (2011) Skeletal muscle-specific expression of PGC-1α-b, an exercise-responsive isoform, increases exercise capacity and peak oxygen uptake. PLoS One 6:e28290
Kok BP, Dyck JR, Harris TE, Brindley DN (2013) Differential regulation of the expressions of the PGC-1α splice variants, lipins, and PPARα in heart compared to liver. J Lipid Res 54:1662–1677
Felder TK, Soyal SM, Oberkofler H et al (2011) Characterization of novel peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) isoform in human liver. J Biol Chem 286:42923–42936
Soyal SM, Felder TK, Auer S et al (2012) A greatly extended PPARGC1A genomic locus encodes several new brain-specific isoforms and influences Huntington disease age of onset. Hum Mol Genet 21:3461–3473
Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D (2006) Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127:59–69
Weydt P, Pineda VV, Torrence AE et al (2006) Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1α in Huntington’s disease neurodegeneration. Cell Metab 4:349–362
Weydt P, Soyal SM, Gellera C et al (2009) The gene coding for PGC-1α modifies age at onset in Huntington’s disease. Mol Neurodegener 4:3
Zhang Y, Huypens P, Adamson AW et al (2009) Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1α. J Biol Chem 284:32813–32826
Chang JS, Fernand V, Zhang Y et al (2012) NT-PGC-1α protein is sufficient to link β3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis. J Biol Chem 287:9100–9111
Wen X, Wu J, Chang JS et al (2014) Effect of exercise intensity on isoform-specific expressions of NT-PGC-1 α mRNA in mouse skeletal muscle. Biomed Res Int 2014:402175
Wu Z, Puigserver P, Andersson U et al (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124
Jun HJ, Gettys TW, Chang JS (2012) Transcriptional activity of PGC-1α and NT-PGC-1α Is differentially regulated by Twist-1 in brown fat metabolism. PPAR Res 2012:320454
Pan D, Fujimoto M, Lopes A, Wang YX (2009) Twist-1 is a PPARδ-inducible, negative-feedback regulator of PGC-1α in brown fat metabolism. Cell 137:73–86
Thom R, Rowe GC, Jang C, Safdar A, Arany Z (2014) Hypoxic induction of vascular endothelial growth factor (VEGF) and angiogenesis in muscle by truncated peroxisome proliferator-activated receptor gamma coactivator (PGC)-1α. J Biol Chem 289:8810–8817
Bostrom P, Wu J, Jedrychowski MP et al (2012) A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481:463–468
Roberts LD, Bostrom P, O’Sullivan JF et al (2014) β-Aminoisobutyric acid induces browning of white fat and hepatic beta-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab 19:96–108
Shan T, Liang X, Bi P, Kuang S (2013) Myostatin knockout drives browning of white adipose tissue through activating the AMPK-PGC1α-Fndc5 pathway in muscle. FASEB J 27:1981–1989
Rao RR, Long JZ, White JP et al (2014) Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell 157:1279–1291
Agudelo LZ, Femenia T, Orhan F et al (2014) Skeletal muscle PGC-1α1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell 159:33–45
Tadaishi M, Miura S, Kai Y et al (2011) Effect of exercise intensity and AICAR on isoform-specific expressions of murine skeletal muscle PGC-1α mRNA: a role of β2-adrenergic receptor activation. Am J Physiol Endocrinol Metab 300:E341–E349
White JP, Wrann CD, Rao RR et al (2014) G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc Natl Acad Sci U S A 111:15756–15761
Kim SH, Asaka M, Higashida K, Takahashi Y, Holloszy JO, Han DH (2013) β-Adrenergic stimulation does not activate p38 MAP kinase or induce PGC-1α in skeletal muscle. Am J Physiol Endocrinol Metab 304:E844–E852
Ydfors M, Fischer H, Mascher H, Blomstrand E, Norrbom J, Gustafsson T (2013) The truncated splice variants, NT-PGC-1α and PGC-1α4, increase with both endurance and resistance exercise in human skeletal muscle. Physiol Rep 1:e00140
Lundberg TR, Fernandez-Gonzalo R, Norrbom J, Fischer H, Tesch PA, Gustafsson T (2014) Truncated splice variant PGC-1α4 is not associated with exercise-induced human muscle hypertrophy. Acta Physiol (Oxf) 212:142–151
Hanson AM, Stodieck LS, Cannon CM, Simske SJ, Ferguson VL (2010) Seven days of muscle re-loading and voluntary wheel running following hindlimb suspension in mice restores running performance, muscle morphology and metrics of fatigue but not muscle strength. J Muscle Res Cell Motil 31:141–153
Perez-Schindler J, Summermatter S, Santos G, Zorzato F, Handschin C (2013) The transcriptional coactivator PGC-1α is dispensable for chronic overload-induced skeletal muscle hypertrophy and metabolic remodeling. Proc Natl Acad Sci U S A 110:20314–20319
Acknowledgements
We thank I. Cervenka (Karolinska Institutet, Stockholm, Sweden) for generating illustrations used in this review and all members of the laboratory for insightful discussions.
Funding
This work was supported by grants from the Swedish Research Council, The Novo Nordisk Foundation (Denmark), The Malin and Lennart Philipson Foundation (Sweden), The Swedish Diabetes Research Foundation, and a Marie Curie Career Integration grant (EU) to JLR. ATP is supported by a postdoctoral fellowship from the Swedish Society for Medical Research (SSMF).
Contribution statement
All authors were responsible for drafting the article and revising it critically for important intellectual content. All authors approved the version to be published.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martínez-Redondo, V., Pettersson, A.T. & Ruas, J.L. The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia 58, 1969–1977 (2015). https://doi.org/10.1007/s00125-015-3671-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-015-3671-z