Abstract
Aims/hypothesis
Chronic foot ulceration is a severe complication of diabetes, driving morbidity and mortality. The mechanisms underlying delaying wound healing in diabetes are incompletely understood and tools to identify such pathways are eagerly awaited.
Methods
Wound biopsies were obtained from 75 patients with diabetic foot ulcers. Matched subgroups of rapidly healing (RH, n = 17) and non-healing (NH, n = 11) patients were selected. Proteomic analysis was performed by labelling with isobaric tag for relative and absolute quantification and mass spectrometry. Differentially expressed proteins were analysed in NH vs RH for identification of pathogenic pathways. Individual sample gene/protein validation and in vivo validation of candidate pathways in mouse models were carried out.
Results
Pathway analyses were conducted on 92/286 proteins that were differentially expressed in NH vs RH. The following pathways were enriched in NH vs RH patients: apoptosis, protease inhibitors, epithelial differentiation, serine endopeptidase activity, coagulation and regulation of defence response. SerpinB3 was strongly upregulated in RH vs NH wounds, validated as protein and mRNA in individual samples. To test the relevance of serpinB3 in vivo, we used a transgenic mouse model with α1-antitrypsin promoter-driven overexpression of human SERPINB3. In this model, wound healing was unaffected by SERPINB3 overexpression in non-diabetic or diabetic mice with or without hindlimb ischaemia. In an independent validation cohort of 47 patients, high serpinB3 protein content was confirmed as a biomarker of healing improvement.
Conclusions/interpretation
We provide a benchmark for the unbiased discovery of novel molecular targets and biomarkers of impaired diabetic wound healing. High serpinB3 protein content was found to be a biomarker of successful healing in diabetic patients.
Similar content being viewed by others
Introduction
Diabetes is burdened by severe multi-organ complications that collectively reduce life expectancy [1]. Foot ulcers are among the most disabling complications, often leading to amputation and excess mortality. Diabetes is indeed the most common cause of non-traumatic limb amputation in western countries [2, 3]. The molecular mechanisms leading to impaired wound healing in diabetes are incompletely understood [4]. Wound healing is a complex process involving several tissues, cell types and biological pathways in a multi-step and highly coordinated series of events [5]. Coagulation, epithelial gap closure, formation and regression of the granulation tissue, angiogenesis and inflammation and its resolution are all prominently involved in wound healing [6]. Alterations in each of the aforementioned events can delay healing and promote complications of the wound. In diabetic patients, foot ulcers result from a combination of neuropathy and ischaemia, are often triggered by traumatic events and are complicated by infection [7]. So far, studies on the mechanisms of impaired wound healing in diabetes have failed to transfer pre-clinical findings into clinical-grade therapeutic strategies. In addition, there is a paucity of translational studies aimed at replicating in humans findings obtained in animal models and vice-versa.
Omics techniques (genomics, proteomics and metabolomics) provide an unbiased opportunity to discover new potential pathogenic mechanisms and therapeutic targets, as well as biomarkers of disease [8]. Such approaches are used to generate hypotheses that need to be verified in animal models and in humans [9]. So far, proteomics has been rarely used to discover new aspects of the delayed diabetic wound healing process, and the resulting pathways have never been validated [10–12]. Furthermore, cross-sectional comparisons of diabetic vs non-diabetic wound tissues have been biased by patients’ differences and underlying conditions [10, 11]. Rather, comparing diabetic patients in whom healing has been followed-up over time will likely provide more reliable data on the molecular mechanisms underlying the healing process.
In this reverse translational study, we first used a gold-standard proteomic approach [13] to characterise the molecular pathways enriched in a discovery cohort of patients with rapidly healing compared with NH wounds. We then selected serpinB3 as a candidate target to be further evaluated in a mouse model and in a clinical validation cohort. We describe a process whereby candidate molecular targets can be identified, validated and distinguished from bystander biomarkers.
Methods
Patients
The study was approved by local Ethical committee and conducted in accordance with the principles of the Declaration of Helsinki as revised in 2000. Patients were recruited at the Diabetic Foot and Ankle Clinic of the Abano Terme Hospital during a 6 month accrual period (from November 2009 to May 2010). All consecutive patients with type 1 or type 2 diabetes aged 18–90 years admitted to the Clinic were considered eligible pending satisfaction of inclusion/exclusion criteria and provision of informed consent. Exclusion criteria were as follows: systemic infection or sepsis; immunosuppression or immunological disorders; inability to provide informed consent; pregnancy or lactation. Patients with neuropathic, ischaemic or neuroischaemic wounds could be enrolled. We collected data on age, sex, BMI, waist circumference, HbA1c, diabetes duration, prevalence of hypertension (blood pressure >140/90 mmHg [systolic/diastolic] on at least two occasions or use of antihypertensive medications), dyslipidaemia (total cholesterol >5.17 mmol/l [200 mg/dl] or LDL-cholesterol >3.36 mmol/l [130 mg/dl] or triacylglycerol >1.69 mmol/l [150 mg/dl]), cigarette smoking (one or more cigarette per day), coronary artery disease (CAD; history of myocardial infarction or angina or evidence of >50% coronary stenosis at a coronary angiography), cerebrovascular disease (history of stroke or transient ischaemic attack or evidence of >50% stenosis of carotid arteries), peripheral vascular disease (instrumental evidence of significant stenosis or leg arteries), retinopathy (defined by ophthalmological examination), neuropathy (based on history, clinical examination and eventually confirmed by electromyography), chronic renal failure (estimated glomerular filtration rate <60 ml min−1 [1.73 m]−2). We also recorded antihyperglycaemic and other medications. Foot ulcers were classified as neuropathic based on the following criteria: typical ulcer site, absence of ischaemia and normal transcutaneous O2 pressure; ischaemia in the presence of necrosis/gangrene without clinical and instrumental signs of neuropathy; neuroischaemia in the presence of clinical/instrumental evidence of both ischaemia and neuropathy. Wound size was estimated by digital photography and edge reconstruction using ImageJ ver. 1.45 (NIH, Bethesda, MD, USA). A skin biopsy of 5 mm in transversal diameter was taken from the wound margin using a surgical scalpel with care taken to avoid collection of debris, necrotic and fibrin-rich areas. Cases in which the physician felt that a skin biopsy could adversely affect wound healing were excluded. Biopsies were snap frozen until biochemical analyses.
Patients were followed up for 6 months to evaluate wound healing and the following endpoints: surgical or endovascular revascularisation; need for surgical debridement; development of new ulcers; minor and major amputations and death.
For definition of the discovery cohort, out of the initial sample of 75 patients undergoing skin biopsy, we selected patients who could be distinguished as rapidly healing (RH; if the healing process was successfully completed within 6 months) or non-healing (NH; if the ulcer persisted or was even enlarged at the end of observation). Each NH patient was matched with one or two RH patients of the same sex and within a ± 5 year difference in age.
The remaining 47 patients were included in the validation cohort and were divided into those showing improvement in wound healing at 6 months (reduction in wound size; healing) and those showing no improvement or worsening of the healing process at 6 months (no change; enlargement of the wound size).
Tissue processing
Biopsies of the wound area were frozen and stored at −80°C. Full-thickness radial slices of the tissue were stored for subsequent independent analysis of protein content and RNA quantification by real-time PCR. Details on proteomic analysis can be found in the electronic supplementary material (ESM) Methods.
Pathway analysis
Bioinformatic analysis was carried out using DAVID Bioinformatics Resources 6.7 (database for annotation, visualization, and integrated discovery; http://david.abcc.ncifcrf.gov/, accessed on 10 October 2013) [14], which relies on a modified Fisher’s exact p value to cluster proteins in functional annotation groups, establishing whether a group is over-represented in a given proteomic data set.
ELISA
Frozen tissue was minced and manually ground with liquid nitrogen. RIPA buffer (Sigma-Aldrich, St Louis, MO, USA) was added and the tissue solution was further disrupted using TissueLyser LT (Qiagen, Venlo, the Netherlands). The solution was centrifuged at 20,000 g for 20 min at 4°C to pellet insoluble proteins and organelles. The supernatant fraction, containing soluble proteins, was collected and centrifuged at 12,000 g for 15 min at 15°C to avoid salt crystallisation. Protein content was quantified using the BCA assay (Thermo Fisher Scientific, Rockford, IL USA). A custom ELISA was used to measure serpinB3 content in tissue extracts. Briefly, 100 μl/well of mouse anti-human serpinB3 monoclonal antibody 8H11 (Santa Cruz Biotechnology, Dallas, TX, USA) diluted in PBS (pH 7.4) at a concentration of 5 μg/ml, was coated on microtitre plates and incubated overnight at 4°C. The wells were blocked with 5% milk powder and incubated at room temperature for 2 h. After washing with PBS-0.05% Tween, 100 μl of each sample was added to plate wells and incubated for 1 h at room temperature. A standard curve was obtained using 16–0.25 ng/ml recombinant serpinB3 [15]. Samples and standards were tested in duplicate. SerpinB3 was revealed by incubating with 100 μl of horseradish peroxidase-conjugated streptavidin secondary anti-serpinB3 antibody. The plate was developed with a ready-to-use TMB substrate solution (Thermo Fisher Scientific). The reaction was stopped with 1 mol/l HCl (100 μl) and absorbance at 450 nm, was measured on a microplate reader (Tecan, San Jose, CA, USA).
Gene expression analysis
Total RNA was extracted using the RNeasy kit (Qiagen) and RNA was reverse transcribed to generate cDNA using the First-Strand cDNA Synthesis Kit from Invitrogen (Carlsbad, CA, USA). Real-time PCR with Fast SYBR Green detection was performed using an 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and the following primers: β-actin forward 5′-AGAGCTACGAGCTGCCTGAC-3′ reverse 5′-GGATGCCACAGGACTCCA-3′; SERPINB3 forward 5′-AGCTATGACCTCAAGGACAC-3′ reverse 5′-ACTCCAGATAGCACGAGAC-3′. Expression data were normalised to the mean of the housekeeping gene β-actin to control the variability in expression levels and were analysed using the \( {2}^{-\Delta {\mathrm{C}}_{\mathrm{t}}} \) method.
Animal models
Experiments involving animals were approved by the local Ethical committee and conducted in accordance with the ‘Principles of laboratory animal care’ (NIH publication no. 85–23, revised 1985; http://grants1.nih.gov/grants/olaw/references/phspol.htm, accessed 5 February 2012). C57Bl/6 mice aged 8–12 weeks were used (either wild-type [WT] or transgenic for human SERPINB3, under the α1-antitripsin promoter [A1at-SerpinB3TG]). A1at-SerpinB3TG mice were generated as previously described [16]. Diabetes was induced by a single intraperitoneal injection of 150 mg/kg streptozotocin (STZ) (Sigma Aldrich) in citrate buffer 50 mmol/l, pH 4.5. Blood glucose was measured with a Glucocard G-meter (Menarini, Florence, Italy); mice with blood glucose ≥ 16.7 mmol/l (300 mg/dl) in at least two measurements within the first week were classified as diabetic and housed for 4 weeks with free access to food and water before performing experiments. The hindlimb wound model was used as previously described [17]. For induction of hindlimb ischaemia, mice were sedated with 10 mg/kg zolazepam/thylamine (Zoletil; Laboratories Virbac, Nice, France) and 7 mg/kg xylazine (Xilor; Bio98, Milan, Italy). The femoral artery and vein were surgically dissected from the femoral nerve, then cauterised with low-temperature cautery and excised between the inguinal ligament and ankle. To create skin wounds, mice were sedated with inhaled sodium isoflurane and the skin of the dorsal surface of the hindlimb was shaved with Veet cream and disinfected with alcohol/ether. Then, a 4 mm diameter wound was performed with a biopsy punch (H-S Medical, Colton, CA, USA). Daily photographs were taken to evaluate wound closure time and the wound area was quantified with ImageJ. Wounds on ischaemic mice were performed 2 days after surgery.
Statistical analysis
Data are presented as mean ± SE for continuous variables, or as percentage for categorical variables. Normality of the variables of interest was assessed using the Kolmogorov–Smirnov test. Comparison between two groups was performed using Student’s t test for normal variables and the Mann–Whitney test for non-normal variables. Categorical variables were compared using the χ 2 test. A multivariable analysis was run to adjust for potential confounders in the association between candidate biomarker(s) and clinical variables or outcomes. A receiver operating characteristic (ROC) curve was constructed to detect the ability of serpinB3 levels to discriminate between patients based on follow-up evaluation of the wound. The optimal cut-off was chosen as that maximising the sum of sensitivity and specificity. Statistical significance was accepted at p < 0.05. SPSS ver. 20.0 (IBM, Armonk, NY, USA) was used.
Results
Proteomic analysis in the discovery cohort
The discovery cohort was composed of age- and sex-matched patients, who could be distinguished clearly into one of two groups: RH patients whose wounds had healed at 6 months and NH patients whose wounds had worsened at 6 months or who had undergone amputation. This clear-cut separation was designed to optimise the chance of finding biomarkers or pathogenic factors of the wound healing process. Clinical characteristics of the discovery cohort are reported in Table 1. Besides expected differences in the outcome, there were no other significant differences between the RH and NH group in concomitant risk factors, diabetes duration and control, complications and therapy. The prevalence of wound infection was also comparable between the two groups. A quantitative proteomic analysis of pools of wound biopsy extracts from the RH and NH groups using isobaric tag for relative and absolute quantification (iTRAQ) identified 286 proteins present in both pools. Using the standard 1.5-fold threshold for upregulation and the 0.6-fold threshold for downregulation, along with the α = 0.05 significance based on the technical replicates, 92 proteins (32.1% of total) were found to be differentially expressed in RH vs NH (ESM Table 1).
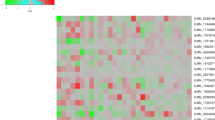
A pathway analysis was performed using the DAVID platform. Biological pathways significantly enriched in the group of differentially expressed proteins over the background of all identified proteins were as follows: apoptosis, protease inhibitor activity, serine-type endopeptidase activity, epithelial cell differentiation, coagulation and negative regulation of defence response (Fig. 1). This analysis shows a net separation of pathways in relation to the wound outcome, with NH wounds having a higher content of proteins related to inhibition of apoptosis, serine endopeptidase activity, coagulation and negative regulation of defence response, concomitantly with a reduction in proteins related to apoptosis induction and epithelial cell differentiation.
Pathways enriched in proteins differentially expressed in RH and NH wound patients. Molecular pathways are indicated on the left, while each column indicates a protein of a pathway found to be differentially expressed in RH vs NH wound patients. Bars indicate error of the internal technical replicates. Circles, pro-apoptotic protein; crosses, anti-apoptotic proteins; Isof., isoform
Validation of serpinB3 in the discovery cohort
Among the proteins that were found to be differentially expressed in wound extracts of the RH vs NH group, we focused on serpinB3, which was 0.55-fold lower in NH vs RH pools (or 1.83-fold higher in RH vs NH pools), based on iTRAQ analysis. We used a custom ELISA to validate serpinB3 concentrations in individual samples of the RH and NH groups. SerpinB3 concentrations were normalised to the total protein concentration to account for potential differences in the composition of each wound biopsy. The serpinB3/protein ratio was twofold higher in RH compared with NH wound extracts, although this difference did not achieve statistical significance, owing to large variability (Fig. 2a). SERPINB3 expression was, however, significantly higher in RH than in NH individual samples (Fig. 2b). As the serpinB3 proteomic probes might not unequivocally distinguish between serpinB3 and serpinB4, we also measured SERPINB4 mRNA expression, which was found to not significantly differ between RH and NH patients (Fig. 2c).
Wound healing in Serpinb3 transgenic mice
As SERPINB3 expression was found to be higher in RH human diabetic wounds, we wondered whether Serpinb3 overexpression in diabetic mice with or without hindlimb ischaemia would affect the wound healing process. To this end, we used C57Bl/6 mice, either WT or A1at-SerpinB3TG. First, we found that the human transgene was expressed in the skin of A1at-SerpinB3TG while, as expected, it was undetectable in WT mice (Fig. 3a). A1at-SerpinB3TG mice also showed a mild and non-significant increase in murine Serpinb3 gene expression (Fig. 3b). These data indicate that the A1at-SerpinB3TG mouse is a good model with which to study the effects of serpinB3 on wound healing. Diabetes was induced in mice by STZ injection and hindlimb ischaemia was induced by ligation and excision of the femoral artery. Healing of a hindlimb 4 mm wound was monitored digitally until closure. Although diabetes and hindlimb ischaemia delayed wound healing as expected, in diabetic and non-diabetic mice with or without hindlimb ischaemia, the time course of wound healing was identical in WT and A1at-SerpinB3TG mice (Fig. 3c–f).
Wound healing in A1at-SerpinB3TG mice. (a) The human SERPINB3 transgene was expressed in the skin of A1at-SerpinB3TG mice, while it was undetectable in WT mice. The real-time PCR amplification plot is also shown. (b) Mouse Serpinb3 gene expression was slightly and not significantly increased in A1at-SerpinB3TG vs WT mice. (c) Wound healing was analysed in WT mice, comparing non-diabetic (circles, solid line), STZ diabetic (squares, dashed line) and STZ diabetic ischaemic (triangles, dotted line). *p < 0.05 comparing STZ diabetic and diabetic ischemic with controls. (d–f) Wound healing was compared in A1at-SerpinB3TG (squares, solid line) vs WT (circles, dashed line) non-diabetic (d), STZ diabetic (e) and STZ diabetic ischaemic (f) mice. While diabetes and ischaemia delayed wound healing in WT mice (c), healing was unaffected in A1at-SerpinB3TG vs WT mice in the presence or absence of diabetes and hindlimb ischaemia (d–f)
Validation of serpinB3 as a biomarker
Although the mouse model suggests that high serpinB3 expression is insufficient to improve diabetic and/or ischaemic wound healing per se, we hypothesised that serpinB3 might nonetheless represent a non-pathogenic biomarker of successful wound healing. Therefore, we measured serpinb3 gene expression and protein content (as the ratio to the total protein concentration) in wound extracts from the validation cohort. Clinical characteristics of this cohort of patients are reported in Table 1. Out of 47 patients, 16 showed wound healing improvement, while 31 showed no improvement or worsened at 6 months. Among all clinical variables, only the prevalence of CAD was significantly different (higher) in patients showing no improvement at follow-up. Importantly, the prevalence of infection was comparable between the two groups. The serpinB3/total protein ratio was 2.12-fold higher in patients who improved than in those who did not (Fig. 4a), while the levels of SERPINB3 and SERPINB4 expression were not significantly different (Fig. 4b, c). There were no differences in serpinB3/total protein ratio between the two groups in relation to the prevalence of CAD, suggesting that CAD was not a confounder in the comparison (not shown). Indeed, in a multivariable analysis adjusted for CAD, which was the only significantly different variable between groups, the serpinB3/total protein ratio remained significantly directly correlated to wound healing improvement (B = 1.26 ± 0.62; β = 0.307; p = 0.049). There was no significant association between serpinB3/total protein ratio or gene expression in relation to the prevalence of infection and other breakdown outcomes, such as amputations, relapse, need for further surgical debridement or death. According to the ROC curve for the serpinB3 protein biomarker of improved wound healing, AUC was 0.665 (95% CI 0.500, 0.831), with sensitivity 75.0% and specificity 62.5% for the optimal cut-off of 1.13 ng ml−1 μg−1 μl−1 (Fig. 4d).
Validation of serpinB3 as a biomarker of wound healing. (a–c) Expression of serpinB3 protein (a) and SERPINB3 (b) and SERPINB4 genes (c) was determined in samples of the validation cohort (see Table 1). *p < 0.05, improved vs not improved patients. (d) A ROC curve was constructed to quantify the capacity of serpinB3 protein concentration above 1.13 ng ml−1 μg−1 μl−1 (optimal cut-off) to distinguish patients in whom the wound healing process improved from those did not improve at follow-up
Discussion
We describe a benchmark for the identification and validation of pathogenic and non-pathogenic biomarkers of impaired diabetic wound healing, based on proteomic analysis of the wound tissue, followed by gene and protein expression confirmation. The use of appropriate animal models is essential to validate candidate pathogenic molecular targets, whereas the association of healing or non-healing with candidate non-pathogenic biomarkers needs to be validated in an independent replication cohort. Using this comprehensive approach, we show that high tissue serpinB3 content is a biomarker of successful wound healing in diabetic patients.
Proteomic analysis of human diabetic wounds has been previously shown to yield reliable and biologically sound information on the molecular mechanisms that might be involved in delayed healing [10, 11]. However, though proteomics is considered to be the way forward in diabetic foot research [12], none of the studies have developed a rigorous validation strategy. In fact, hypothesis-generating omics approaches have been criticised for being burdened by excess rates of false-positive results [18, 19], thus limiting the chances of finding true biomarkers or pathogenic factors. We herein used a gold-standard technique for the simultaneous identification and quantification of proteins in wound biopsies obtained from well-characterised diabetic patients, who could be unequivocally categorised as ‘rapidly healing’ or ‘non-healing’, based on follow-up visits. This clinical approach is critical to define the optimal discovery cohort and maximises the chance of identifying proteins associated with healing efficiency. A bioinformatic analysis selected pathways enriched in proteins that were differentially expressed between the RH and NH groups. Interestingly, all such pathways are plausibly associated with the healing process, being related to coagulation, epithelial integrity, cell survival and extracellular matrix remodelling.
We have focused on serpinB3, as one of the candidate molecular targets. SerpinB3 belongs to the family of evolutionary conserved serine protease inhibitors, the members of which exert a wide range of biological activity. Although the physiological functions of serpinB3 remain elusive, they traditionally include links to cell death and immunity. SerpinB3 is produced by epithelial cells, endothelium and blood cells and its expression is induced by TNF-α. [20]. It has been previously shown to be involved in wound healing processes, including epithelial-to-mesenchymal transition [21], inflammation [22] and apoptosis [23]. Proteomic data, as well as gene and protein analysis, showed higher levels of serpinB3 in RH compared with NH wounds. To evaluate whether serpinB3 can itself promote wound healing, we studied healing kinetics of hindlimb wounds in WT mice and in mice overexpressing human SERPINB3 (A1at-SerpinB3TG). The transgene was expressed in the skin at a level similar to that of the mouse Serpinb3 gene, making this model suitable to test the effects of serpinB3 in wound healing. The hindlimb wound model has the advantage of resembling human disease, because healing is significantly impaired by diabetes and hindlimb ischaemia. We have previously validated this model by showing that quenching oxidative stress by p66Shc deletion promotes healing of diabetic and ischaemic wounds [17]. Despite A1at-SerpinB3TG mice having several pathological and non-pathological features comparable with those of WT mice [21–23], including an increased lifespan [24], we did not find any difference in the wound healing process. This indicates that, at least in this experimental model, a high serpinB3 level is in itself insufficient to improve wound healing and seems to argue against a direct role for serpinB3 in the healing process. SerpinB3 normally inhibits the activity of proteases (such as papain and cathepsins), many of which are associated with the healing process and may themselves represent pathogenic mechanisms (Fig. 1). In fact, an imbalance between metalloproteases and their inhibitors appears be associated with features of diabetic wounds [25, 26]. However, the transgenic expression of the human serpinB3 protein may be insufficient to detect differences in the healing process compared with the control condition, because control mice have physiological serpinB3 levels and activity. Further data on the role played by serpinB3 in experimental wound healing may arise from the analysis of Serpinb3 −/−mice.
Notwithstanding such limitations, serpinB3 levels may serve as a biomarker of wound healing in diabetic patients. To test this hypothesis, we determined serpinB3 protein and gene expression in wound biopsies from patients in the validation cohort, who were followed-up for 6 months to collect data on wound outcomes and death. Interestingly, serpinB3 protein levels were on average higher in patients who showed an improvement in wound healing at follow-up compared with those who showed no improvement, while gene expression did not differ. This discrepancy may be explained by differences in protein vs mRNA stability [27], or it may suggest that the high level of serpinB3 protein in healing patients is produced systemically and not locally by wound cells. The latter hypothesis may be verified by measuring plasma/serum serpinB3 concentrations in patients with foot ulcers. For instance, patients with psoriasis have elevated serum serpinB3 levels in relation to disease activity [28]. It should be noted that while the discovery cohort in the current study was artificially created by matching patients clearly corresponding to healing and NH phenotypes, the validation cohort was composed of unselected patients and is more similar to diabetic patients with foot ulcers seen in routine clinical practice. Based on the ROC curve, the relative serpinB3 protein content showed the ability to discriminate between patients who showed signs of wound improvement and those who did not improve at follow-up. Accuracy, measured as AUC, was relatively low, likely due to the dispersion of wound protein content of serpinB3. As serpinB3 can reflect features of the ongoing systemic inflammatory process, measurement of serum serpinB3 concentrations may provide a more accurate discrimination between healing and non-healing patients. In addition to standard clinical assessment, this biomarker could be used to drive therapeutic decisions needed to improve wound and patient outcomes.
In conclusion, we have designed and applied for the first time a benchmark to identify and validate pathogenic and non-pathogenic biomarkers of wound healing in diabetic patients (Fig. 5). We show how this process led us to identify serpinB3 as a biomarker of successful healing, which could be employed clinically to stratify the probability of good vs bad wound outcome. The same pre-clinical and clinical validation should be used for other candidate molecular targets arising from the unbiased proteomic signature of wound healing we describe: regulators of apoptosis and extracellular matrix remodelling are emerging targets to be explored.
Abbreviations
- CAD:
-
Coronary artery disease
- iTRAQ:
-
Isobaric tag for relative and absolute quantification
- NH:
-
Non-healing
- RH:
-
Rapidly healing
- STZ:
-
Streptozotocin
- WT:
-
Wild-type
References
Seshasai SR, Kaptoge S, Thompson A et al (2011) Diabetes mellitus, fasting glucose, and risk of cause-specific death. N Engl J Med 364:829–841
Morbach S, Furchert H, Groblinghoff U et al (2012) Long-term prognosis of diabetic foot patients and their limbs: amputation and death over the course of a decade. Diabetes Care 35:2021–2027
Schaper NC, Apelqvist J, Bakker K (2012) Reducing lower leg amputations in diabetes: a challenge for patients, healthcare providers and the healthcare system. Diabetologia 55:1869–1872
Gary Sibbald R, Woo KY (2008) The biology of chronic foot ulcers in persons with diabetes. Diabetes Metab Res Rev 24(Suppl 1):S25–S30
Clark RA (1985) Cutaneous tissue repair: basic biologic considerations. I. J Am Acad Dermatol 13:701–725
Broughton G 2nd, Janis JE, Attinger CE (2006) The basic science of wound healing. Plast Reconstr Surg 117:12S–34S
American Diabetes Association (1999) Consensus Development Conference on Diabetic Foot Wound Care: 7-8 April 1999, Boston, Massachusetts. Diabetes Care 22:1354–1360
Kussmann M, Raymond F, Affolter M (2006) OMICS-driven biomarker discovery in nutrition and health. J Biotechnol 124:758–787
Gjesing AP, Pedersen O (2012) ‘Omics’-driven discoveries in prevention and treatment of type 2 diabetes. Eur J Clin Invest 42:579–588
Eming SA, Koch M, Krieger A et al (2010) Differential proteomic analysis distinguishes tissue repair biomarker signatures in wound exudates obtained from normal healing and chronic wounds. J Proteome Res 9:4758–4766
Krisp C, Jacobsen F, McKay MJ et al (2013) Proteome analysis reveals antiangiogenic environments in chronic wounds of diabetes mellitus type 2 patients. Proteomics 13:2670–2681
Sweitzer SM, Fann SA, Borg TK, Baynes JW, Yost MJ (2006) What is the future of diabetic wound care? Diabetes Educ 32:197–210
Ross PL, Huang YN, Marchese JN et al (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3:1154–1169
da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Turato C, Biasiolo A, Pengo P et al (2011) Increased antiprotease activity of the SERPINB3 polymorphic variant SCCA-PD. Exp Biol Med (Maywood) 236:281–290
Villano G, Quarta S, Ruvoletto MG et al (2010) Role of squamous cell carcinoma antigen-1 on liver cells after partial hepatectomy in transgenic mice. Int J Mol Med 25:137–143
Fadini GP, Albiero M, Menegazzo L et al (2010) The redox enzyme p66Shc contributes to diabetes and ischemia-induced delay in cutaneous wound healing. Diabetes 59:2306–2314
Mehta TS, Zakharkin SO, Gadbury GL, Allison DB (2006) Epistemological issues in omics and high-dimensional biology: give the people what they want. Physiol Genomics 28:24–32
Kirk P, Witkover A, Bangham CR et al (2013) Balancing the robustness and predictive performance of biomarkers. J Comput Biol 20:979–989
Gatto M, Iaccarino L, Ghirardello A et al (2013) Serpins, immunity and autoimmunity: old molecules, new functions. Clin Rev Allergy Immunol 45:267–280
Quarta S, Vidalino L, Turato C et al (2010) SERPINB3 induces epithelial-mesenchymal transition. J Pathol 221:343–356
Villano G, Lunardi F, Turato C et al (2012) Increased Th1 immune response in SERPINB3 transgenic mice during acute liver failure. Exp Biol Med (Maywood) 237:1474–1482
Vidalino L, Doria A, Quarta S et al (2009) SERPINB3, apoptosis and autoimmunity. Autoimmun Rev 9:108–112
Villano G, Ruvoletto M, Ceolotto G et al (2013) SERPINB3 is associated with longer survival in transgenic mice. Sci Rep 3:3056
Dinh T, Tecilazich F, Kafanas A et al (2012) Mechanisms involved in the development and healing of diabetic foot ulceration. Diabetes 61:2937–2947
Menghini R, Uccioli L, Vainieri E et al (2013) Expression of tissue inhibitor of metalloprotease 3 is reduced in ischemic but not neuropathic ulcers from patients with type 2 diabetes mellitus. Acta Diabetol 50:907–910
Schwanhausser B, Busse D, Li N et al (2011) Global quantification of mammalian gene expression control. Nature 473:337–342
Hamanaka S, Ujihara M, Numa F, Kato H (1997) Serum level of squamous cell carcinoma antigen as a new indicator of disease activity in patients with psoriasis. Arch Dermatol 133:393–395
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
All authors contributed substantially to study conception and design, data acquisition, analysis and interpretation and drafting or revising the manuscript. All authors gave their approval to the final version of the manuscript. GPF is the guarantor of this work.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Methods
(PDF 73 kb)
ESM Table 1
(PDF 137 kb)
Rights and permissions
About this article
Cite this article
Fadini, G.P., Albiero, M., Millioni, R. et al. The molecular signature of impaired diabetic wound healing identifies serpinB3 as a healing biomarker. Diabetologia 57, 1947–1956 (2014). https://doi.org/10.1007/s00125-014-3300-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-014-3300-2