Abstract
Aims/hypothesis
The PPARGC1A gene coactivates multiple nuclear transcription factors involved in cellular energy metabolism and vascular stasis. In the present study, we genotyped 35 tagging polymorphisms to capture all common PPARGC1A nucleotide sequence variations and tested for association with metabolic and cardiovascular traits in 2,101 Danish and Estonian boys and girls from the European Youth Heart Study, a multicentre school-based cross-sectional cohort study.
Methods
Fasting plasma glucose concentrations, anthropometric variables and blood pressure were measured. Habitual physical activity and aerobic fitness were objectively assessed using uniaxial accelerometry and a maximal aerobic exercise stress test on a bicycle ergometer, respectively.
Results
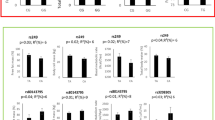
In adjusted models, nominally significant associations were observed for BMI (rs10018239, p = 0.039), waist circumference (rs7656250, p = 0.012; rs8192678 [Gly482Ser], p = 0.015; rs3755863, p = 0.02; rs10018239, beta = −0.01 cm per minor allele copy, p = 0.043), systolic blood pressure (rs2970869, p = 0.018) and fasting glucose concentrations (rs11724368, p = 0.045). Stronger associations were observed for aerobic fitness (rs7656250, p = 0.005; rs13117172, p = 0.008) and fasting glucose concentrations (rs7657071, p = 0.002). None remained significant after correcting for the number of statistical comparisons. We proceeded by testing for gene × physical activity interactions for the polymorphisms that showed nominal evidence of association in the main effect models. None of these tests was statistically significant.
Conclusions/interpretation
Variants at PPARGC1A may influence several metabolic traits in this European paediatric cohort. However, variation at PPARGC1A is unlikely to have a major impact on cardiovascular or metabolic health in these children.
Similar content being viewed by others
Introduction
The protein encoded by the PPARGC1A gene coactivates multiple transcription factors involved in the regulation of oxidative stress, mitochondrial biogenesis, and lipid and glucose oxidation. Exposure to cold, exercise and fasting have all been shown to influence PPARGC1A transcription rates [1], raising the possibility that the gene interacts with energy balance behaviours to influence cardiovascular and metabolic health.
The most frequently studied PPARGC1A polymorphism is the Gly482Ser (rs8192678) non-synonymous coding variant. Associations between Gly482Ser and type 2 diabetes [2, 3], hypertension [4], obesity [5], dyslipidaemia [6–8], aerobic fitness [9] and insulin resistance [10] have been widely reported. However, a thorough exploration of common PPARGC1A variants and their associations with metabolic or cardiovascular disease traits is yet to be reported in adults or children.
The purpose of the present study was to comprehensively assess associations between common PPARGC1A sequence variants and both cardiovascular and metabolic disease traits in Danish and Estonian children from the European Youth Heart Study (EYHS).
Methods
Participants and clinical measures
The EYHS is a multicentre school-based cross-sectional cohort study of European children and adolescents. The methods for the non-genetic aspects of the study have been reported in detail previously [11]. This study comprises 1,255 children (age range boys [8.4–11.2 years] and girls [8.4–11.3 years]) and 846 adolescents (age range: boys [14.1–17.8 years] and girls [14.1–17.0 years]) from Denmark and Estonia (Table 1). Written informed consent was obtained from a parent or guardian and all children gave verbal assent. Local research ethics committees of the respective centres approved the study protocol.
Weight, height and waist circumference were measured using standard anthropometric techniques. BMI was calculated as weight (kg) divided by height (m2) and was standardised to age and sex using the methods described by Cole et al. [12]. Sexual maturity was assessed with the five-stage Tanner scale for breast development in girls and pubic hair in boys [13]. Blood pressure was measured with a paediatric/adult and neonatal vital signs monitor (Dinamap model XL; Critikron, Tampa, FL, USA). Plasma glucose was analysed using the hexokinase method, measured with an autoanalyser (AU600; Olympus Diagnostica, Olympus America, Center Valley, PA, USA). Habitual physical activity was assessed using uniaxial accelerometry (MTI Actigraph, WAM 7164; MTI, Fort Walton Beach, FL, USA) during two weekdays and two weekend days of free-living. The outcome variable in this report is expressed as total counts per unit time (counts/min), which is an indicator of the total volume of activity (i.e. average intensity of activity). Aerobic fitness was assessed during an incremental-load exercise stress test to exhaustion on an electronically braked bicycle ergometer (Monark 839; Ergomedic, Varberg, Sweden). Aerobic fitness is expressed as maximally attained mechanical power relative to body weight (W/kg).
Genetic analyses
To capture all common PPARGC1A sequence variation (±10 kb) we undertook pair-wise tagging (r 2 > 0.8 and minor allele frequency >0.05) using the Tagger software in Haploview (www.broad.mit.edu/mpg/haploview, accessed 1 May 2008) to select 35 tagging polymorphisms from the CEU (Centre d'Etude du Polymorphisme Humain trios originating from northern and western Europe living in Utah, USA) phase 2 HAPMAP (release 19, NCBI build 34) database (for overview of HAPMAP coverage, see Electronic supplementary material [ESM] Table 1). Almost all PPARGC1A genotyping was performed using the Illumina Beadstation custom-array (Illumina, San Diego, CA, USA). The exception was for Gly482Ser (rs8192678), which was genotyped at the MRC Epidemiology Unit genotyping facility using TaqMan (Applied Biosystems, Foster City, CA, USA). A total of 62 DNA samples were of inadequate quality and failed for all assays. Therefore, these were excluded from the dataset. The phenotypic characteristics of these samples did not differ from the remainder of the cohort. The genotyping success rate for all SNPs on the Illumina platform exceeded 98%. The success rate of the TaqMan genotyped Gly482Ser (rs8192678) variant was 96.4%. All polymorphisms were in Hardy–Weinberg equilibrium (p > 0.05), which was assessed using a likelihood ratio test (ESM Table 2). The minor allele frequencies of the polymorphisms ranged from 0.05 to 0.47 (ESM Table 2). Pair-wise correlations between variants ranged from r 2 = 0.01 to r 2 = 0.90.
Statistical power
Calculations were performed using plausible effect sizes (i.e. a beta equivalent to 0.005, 0.01 and 0.015% phenotypic difference per copy of the effective allele) and for a range of minor allele frequencies (0.05–0.45) using QUANTO (V1.2.3) [14] (ESM Fig. 1a–f).
Statistical analysis
Statistical analyses were conducted using SAS software (version 9.1; SAS Institute, Cary, NC, USA). Waist circumference and glucose were logarithmically transformed to normalise their distribution. Generalised linear models were used to test associations between each polymorphism and each outcome trait assuming an additive genetic model. All models were adjusted for country, age, age group, sex and sexual maturity. Adjusting for study centre instead of country did not materially affect the results. Where BP was the dependent variable, adjustments were also made for height and weight. To assess the putative effect modifying roles of age, country or physical activity, interaction terms were fitted to the linear models. Adjustments for multiple statistical comparisons were made using the Holm procedure [15]. Using this approach, the p values for the hypothesis tests are ranked, with the least significant appearing at the top of the list. The denominator for each correction is defined by the number of tests within that rank-ordered list with higher p values. Thus, the least significant hypothesis test is divisible by 1, the second least significant hypothesis test by 2 and so on until all p values are corrected. This correction method was applied to each phenotype separately.
Results
Table 1 shows characteristics of the 1,255 children and 846 adolescents who participated in this study. Significant differences by age group and sex were observed in the proportion of obese individuals within these groups and the levels of metabolic and cardiovascular traits (p < 0.001).
After adjustment, several nominally significant associations were observed for BMI (rs10018239 beta = −0.06 kg/m2 per minor allele copy, p = 0.039), waist circumference (rs10018239 beta = −0.01 cm per minor allele copy, p = 0.043; rs7656250 beta = 0.01 cm per minor allele copy, p = 0.012; rs8192678 [Gly482Ser] beta = 0.01 cm per minor allele copy, p = 0.015; rs3755863 beta = −0.01 cm per minor allele copy, p = 0.02), SBP (rs2970869 beta = 0.77 mmHg per minor allele copy, p = 0.018) and fasting glucose concentrations (rs11724368 beta = −0.01 mmol/l per minor allele copy, p = 0.045). Stronger associations were observed for aerobic fitness (rs7656250 beta = −0.06 W/kg per minor allele copy, p = 0.005; rs13117172 beta = 0.06 W/kg per minor allele copy, p = 0.008) and fasting glucose concentrations (rs7657071 beta = −0.01 mmol/l per minor allele copy, p = 0.002). After correcting for multiple comparisons (each phenotype separately), the statistical significance of all models was abolished; the strongest corrected p value was for fasting glucose (rs7657071, p corrected = 0.07) (see ESM Tables 3 and 4 for summary of nominal tests).
Table 2 shows associations between the Gly482Ser polymorphism and a range of previously studied cardiovascular and metabolic traits. In these models, as reported above, waist circumference was the only nominally associated trait (p = 0.02).
We proceeded by testing interaction terms (gene × physical activity, age or country) for each of the polymorphisms that showed nominal evidence of association in the main effect models. None of these interaction tests approached statistical significance.
Discussion
This is, to our knowledge, the first study to comprehensively examine the role of common PPARGC1A sequence variation in relation to complex disease traits. We observed limited evidence supporting the possibility that variants at PPARGC1A influence metabolic or cardiovascular risk factor levels in this cohort of European children. The strongest associations were for the rs7656250 and rs13117172 variants (with aerobic fitness) and the rs7657071 variant (with fasting glucose concentrations), which in this cohort are in very low linkage disequilibrium (LD) (r 2 = 0.13) (see ESM Fig. 1 for LD plot comparing all SNPs).
PPARGC1A is a strong biological candidate for cardiovascular and metabolic disease. Its well established role as a master regulator of energy metabolism, vascular homeostasis, oxidative stress and adipogenesis has prompted many papers reporting on associations between PPARGC1A variants and a range of seemingly plausibly disease traits. The most widely studied PPARGC1A variant is the Gly482Ser non-synonymous polymorphism. Since the initial report of association between this variant and type 2 diabetes [2], many others have reported statistical associations with diabetes-related traits [3, 5, 7, 9, 10]. Here we observed a nominal statistical association between Gly482Ser and waist circumference, but no associations were observed for other traits. There are several possible explanations for the apparent inconsistency between our findings and those previously reported. For example, most existing studies were conducted in adults of advanced age, from a variety of ethnic backgrounds. By contrast, the cohort studied here was of relatively healthy children of northern European ancestry. Most previous studies report nominally statistically significant findings that are rarely adjusted for multiple statistical comparisons, predisposing some to type 2 error. It is also possible that a degree of publication bias exists, where association studies with statistically significant findings are disproportionally represented in the literature. For example, in a recent meta-analysis of published studies, a statistical association between the Gly482Ser polymorphism and type 2 diabetes was reported [3]. However, this finding was not supported by a meta-analysis of type 2 diabetes genetics conducted in a somewhat larger total sample [16]. Similarly, in a recent meta-analysis of Gly482Ser and BP, statistical associations were evident when including only previously published data, but these effects were abolished when unpublished data were also included [4].
PPARGC1A is a relatively large gene, comprised of eight exons. In European whites, approximately 35 tagging variants are required to capture all common variation across the gene. Thus, a caveat to undertaking a comprehensive study of PPARGC1A is that it involves testing multiple hypotheses. To diminish the possibility of false discovery, we present corrected and uncorrected p values for each nominally significant test. However, it may be that corrections such as these are overly conservative. This, combined with the fact that our study was underpowered for some of the association tests performed (ESM Fig. 2a–f), should be born in mind when considering the relevance of some of the negative findings reported herein.
In summary, we have undertaken the first comprehensive study of PPARGC1A sequence variation. Our findings suggest that PPARGC1A variation may play a relatively modest role in regulating some of the cardiovascular and metabolic traits studied here, with the rs7657071 and rs7656250 variants being the most promising candidates for future exploration.
Abbreviations
- EYHS:
-
European Youth Heart Study
- LD:
-
Linkage disequilibrium
References
Soyal S, Krempler F, Oberkofler H, Patsch W (2006) PGC-1alpha: a potent transcriptional cofactor involved in the pathogenesis of type 2 diabetes. Diabetologia 49:1477–1488
Ek J, Andersen G, Urhammer SA et al (2001) Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia 44:2220–2226
Barroso I, Luan J, Sandhu M et al (2006) Meta-analysis of the Gly482Ser variant in PPARGC1A in type 2 diabetes and related phenotypes. Diabetologia 49:501–505
Vimaleswaran KS, Luan J, Andersen G et al (2008) The Gly482Ser genotype at the PPARGC1A gene and elevated blood pressure: a meta-analysis of 13,949 individuals. J Appl Physiol 105:1352–1358
Ridderstrale M, Johansson LE, Rastam L, Lindblad U (2006) Increased risk of obesity associated with the variant allele of the PPARGC1A Gly482Ser polymorphism in physically inactive elderly men. Diabetologia 49:496–500
Muller YL, Bogardus C, Pedersen O, Baier L (2003) A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes 52:895–898
Franks PW, Ekelund U, Brage S et al (2007) PPARGC1A coding variation may initiate impaired NEFA clearance during glucose challenge. Diabetologia 50:569–573
Vohl MC, Houde A, Lebel S, Hould FS, Marceau P (2005) Effects of the peroxisome proliferator-activated receptor-gamma co-activator-1 Gly482Ser variant on features of the metabolic syndrome. Mol Genet Metab 86:300–306
Lucia A, Gomez-Gallego F, Barroso I et al (2005) PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J Appl Physiol 99:344–348
Fanelli M, Filippi E, Sentinelli F et al (2005) The Gly482Ser missense mutation of the peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha) gene associates with reduced insulin sensitivity in normal and glucose-intolerant obese subjects. Dis Markers 21:175–180
Ekelund U, Anderssen SA, Froberg K, Sardinha LB, Andersen LB, Brage S (2007) Independent associations of physical activity and cardiorespiratory fitness with metabolic risk factors in children: the European youth heart study. Diabetologia 50:1832–1840
Cole TJ, Green PJ (1992) Smoothing reference centile curves: the LMS method and penalized likelihood. Stat Med 11:1305–1319
Tanner JM (1962) Growth at adolescence, with a general consideration of the effects of hereditary and environmental factors upon growth and maturation from birth to maturity, 2nd edn. Blackwell, Oxford
Gauderman WJ, Morrison JM (2006) QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. Available from http://hydra.usc.edu/gxe, accessed 1 May 2008
Holm S (1979) A simple sequentially rejective Bonferroni test procedure. Scand J Stat 6:65–70
Zeggini E, Scott LJ, Saxena R et al (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40:638–645
Acknowledgements
We thank the volunteers who participated in the EYHS. We thank M. Sims and the Technical Team (MRC Epidemiology Unit, Institute of Metabolic Sciences, Cambridge, UK) for expert technical assistance with genotyping. Genetic analyses were supported by the Medical Research Council (UK). The EYHS was supported by the following grants: The Danish Heart Foundation, The Danish Medical Research Council Health Foundation, The Danish Council for Sports Research, The Foundation in Memory of Asta Florida Bolding Renée Andersen, The Faculty of Health Sciences, University of Southern Denmark, The Estonian Science Foundation grant numbers 3277 and 5209, and The Medical Research Council, UK. E. C. Brito received a studentship from the Portuguese Foundation for Science and Technology. This work was supported in part with grants from Västerbottens Health Authority (P. W. Franks: strategic appointment 2006-09), Novo Nordisk (370579201), the Swedish Diabetes Association (DIA2006-013) and the Swedish Heart–Lung Foundation.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
Extent to which the 35 PPARGC1A tagging single nucleotide polymorphisms (tag SNPs) capture the PPARGC1A SNPs available in the latest release of the phase 2 HAPMAP (release 24; NCBI build 36) (PDF 88.4 KB)
ESM Table 2
Minor allele frequencies and p values for tests of Hardy–Weinberg equilibrium for PPARGC1A genotypes stratified by country and for the entire EYHS cohort (PDF 97.9 KB)
ESM Table 3
Results for association tests between PPARGC1A genotypes and measures of body composition and aerobic fitness (PDF 131 KB)
ESM Table 4
Results for association tests between PPARGC1A genotypes and blood pressures and fasting glucose concentrations (PDF 140 KB)
ESM Fig. 1
Pairwise linkage disequilibrium (r2) between PPARGC1A markers in the European Youth Heart Study (n = 2101) (PDF 86.2 KB)
ESM Fig. 2
Statistical power estimates for BMI, waist circumference, aerobic fitness, diastolic and systolic blood pressure and glucose analyses in European Youth Heart Study (n = 2101) (PDF 21.9 KB)
Rights and permissions
About this article
Cite this article
Brito, E.C., Vimaleswaran, K.S., Brage, S. et al. PPARGC1A sequence variation and cardiovascular risk-factor levels: a study of the main genetic effects and gene × environment interactions in children from the European Youth Heart Study. Diabetologia 52, 609–613 (2009). https://doi.org/10.1007/s00125-009-1269-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-009-1269-z