Abstract
Key message
Thirty environmentally stable QTL controlling grain size and/or plant height were identified, among which QTgw.cau-7D was delimited into the physical interval of approximately 4.4 Mb.
Abstract
Grain size and plant height (PHT) are important agronomic traits in wheat breeding. To dissect the genetic basis of these traits, we conducted a quantitative trait locus (QTL) analysis using recombinant inbred lines (RILs). In total, 30 environmentally stable QTL for thousand grain weight (TGW), grain length (GL), grain width (GW) and PHT were detected. Notably, one major pleiotropic QTL on chromosome arm 3DS explained the highest phenotypic variance for TGW, GL and PHT, and two stable QTL (QGw.cau-4B and QGw.cau-7D) on chromosome arms 4BS and 7DS contributed greater effects for GW. Furthermore, the stable QTL controlling grain size (QTgw.cau-7D and QGw.cau-7D) were delimited into the physical interval of approximately 4.4 Mb harboring 56 annotated genes. The elite NILs of QTgw.cau-7D increased TGW by 12.79–21.75% and GW by 4.10–8.47% across all three environments. Collectively, these results provide further insight into the genetic basis of TGW, GL, GW and PHT, and the fine-mapped QTgw.cau-7D will be an attractive target for positional cloning and marker-assisted selection in wheat breeding programs.




Similar content being viewed by others
References
Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320–2325. https://doi.org/10.1038/nprot.2006.384
Basten CJ, Weir BS, Zeng Z-B (1997) QTL cartographer: a reference manual and tutorial for QTL mapping. Department of Statistics, North Carolina State University, Raleigh
Borrill P, Ramirez-Gonzalez R, Uauy C (2016) expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol 170:2172–2186. https://doi.org/10.1104/pp.15.01667
Brinton J, Simmonds J, Minter F, Leverington-Waite M, Snape J, Uauy C (2017) Increased pericarp cell length underlies a major quantitative trait locus for grain weight in hexaploid wheat. New Phytol 215:1026–1038. https://doi.org/10.1111/nph.14624
Cabral AL, Jordan MC, Larson G, Somers DJ, Humphreys DG, McCartney CA (2018) Relationship between QTL for grain shape, grain weight, test weight, milling yield, and plant height in the spring wheat cross RL4452/‘AC Domain’. PLoS ONE 13:e0190681
Carter AH, Garland-Campbell K, Morris CF, Kidwell KK (2012) Chromosomes 3B and 4D are associated with several milling and baking quality traits in a soft white spring wheat (Triticum aestivum L.) population. Theor Appl Genet 124:1079–1096. https://doi.org/10.1007/s00122-011-1770-x
Chai L, Chen Z, Bian R, Zhai H, Cheng X, Peng H, Yao Y, Hu Z, Xin M, Guo W, Sun Q, Zhao A, Ni Z (2018) Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor Appl Genet 131:2621–2637. https://doi.org/10.1007/s00122-018-3177-4
Chen J, Chen G, Li Q, Zhang H, Shi C, Sun C, Deng Z, Liu K, Gu Z, Tian J (2014) Construction of genetic map using genotyping chips and QTL analysis of grain weight. Sci Agric Sin 47:4769–4779. https://doi.org/10.3864/j.issn.0578-1752.2014.24.001
Chen WG, Sun DZ, Yan X, Li RZ, Wang SG, Shi YG, Jing RL (2019) QTL analysis of wheat kernel traits, and genetic effects of qKW-6A on kernel width. Euphytica 215:11. https://doi.org/10.1007/s10681-018-2333-x
Cheng X, Chai L, Chen Z, Xu L, Zhai H, Zhao A, Peng H, Yao Y, You M, Sun Q, Ni Z (2015) Identification and characterization of a high kernel weight mutant induced by gamma radiation in wheat (Triticum aestivum L.). BMC Genet 16:127. https://doi.org/10.1186/s12863-015-0285-x
Cheng R, Kong Z, Zhang L, Xie Q, Jia H, Yu D, Huang Y, Ma Z (2017) Mapping QTLs controlling kernel dimensions in a wheat inter-varietal RIL mapping population. Theor Appl Genet 130:1405–1414. https://doi.org/10.1007/s00122-017-2896-2
Cui F, Zhao C, Ding A, Li J, Wang L, Li X, Bao Y, Li J, Wang H (2014) Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet 127:659–675. https://doi.org/10.1007/s00122-013-2249-8
Cui F, Fan X, Chen M, Zhang N, Zhao C, Zhang W, Han J, Ji J, Zhao X, Yang L, Zhao Z, Tong Y, Wang T, Li J (2016) QTL detection for wheat kernel size and quality and the responses of these traits to low nitrogen stress. Theor Appl Genet 129:469–484. https://doi.org/10.1007/s00122-015-2641-7
Deng Z, Cui Y, Han Q, Fang W, Li J, Tian J (2017) Discovery of consistent QTLs of wheat spike-related traits under nitrogen treatment at different development stages. Front Plant Sci 8:2120. https://doi.org/10.3389/fpls.2017.02120
Dong LL, Wang FM, Liu T, Dong ZY, Li AL, Jing RL, Mao L, Li YW, Liu X, Zhang KP, Wang DW (2014) Natural variation of TaGASR7-A1 affects grain length in common wheat under multiple cultivation conditions. Mol Breeding 34:937–947. https://doi.org/10.1007/s11032-014-0087-2
Dong H, Yan S, Liu J, Liu P, Sun J (2019) TaCOLD1 defines a new regulator of plant height in bread wheat. Plant Biotechnol J 17:687–699. https://doi.org/10.1111/pbi.13008
Duan P, Rao Y, Zeng D, Yang Y, Xu R, Zhang B, Dong G, Qian Q, Li Y (2014) SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J 77:547–557. https://doi.org/10.1111/tpj.12405
Eriksen L, Borum F, Jahoor A (2003) Inheritance and localisation of resistance to Mycosphaerella graminicola causing septoria tritici blotch and plant height in the wheat (Triticum aestivum L.) genome with DNA markers. Theor Appl Genet 107:515–527. https://doi.org/10.1007/s00122-003-1276-2
Flintham JE, Borner A, Worland AJ, Gale MD (1997) Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J Agric Sci 128:11–25. https://doi.org/10.1017/S0021859696003942
Gegas VC, Nazari A, Griffiths S, Simmonds J, Fish L, Orford S, Sayers L, Doonan JH, Snape JW (2010) A genetic framework for grain size and shape variation in wheat. Plant Cell 22:1046–1056. https://doi.org/10.1105/tpc.110.074153
Guan P, Lu L, Jia L, Kabir MR, Zhang J, Lan T, Zhao Y, Xin M, Hu Z, Yao Y, Ni Z, Sun Q, Peng H (2018) Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front Plant Sci 9:529. https://doi.org/10.3389/fpls.2018.00529
Guan PF, Di N, Mu Q, Shen XY, Wang YF, Wang XB, Yu KH, Song WJ, Chen YM, Xin MM, Hu ZR, Guo WL, Yao YY, Ni ZF, Sun QX, Peng HR (2019) Use of near-isogenic lines to precisely map and validate a major QTL for grain weight on chromosome 4AL in bread wheat (Triticum aestivum L.). Theor Appl Genet 132:2367–2379. https://doi.org/10.1007/s00122-019-03359-4
Guo T, Chen K, Dong NQ, Shi CL, Ye WW, Gao JP, Shan JX, Lin HX (2018) GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30:871–888. https://doi.org/10.1105/tpc.17.00959
Hanif M, Gao F, Liu J, Wen W, Zhang Y, Rasheed A, Xia X, He Z, Cao S (2015) TaTGW6-A1, an ortholog of rice TGW6, is associated with grain weight and yield in bread wheat. Mol Breeding 36:1. https://doi.org/10.1007/s11032-015-0425-z
Huang XQ, Coster H, Ganal MW, Roder MS (2003) Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor Appl Genet 106:1379–1389. https://doi.org/10.1007/s00122-002-1179-7
Huang Y, Kong Z, Wu X, Cheng R, Yu D, Ma Z (2015) Characterization of three wheat grain weight QTLs that differentially affect kernel dimensions. Theor Appl Genet 128:2437–2445. https://doi.org/10.1007/s00122-015-2598-6
International Wheat Genome Sequencing C (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. https://doi.org/10.1126/science.1251788
International Wheat Genome Sequencing C (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. https://doi.org/10.1126/science.aar7191
Jiang Y, Jiang Q, Hao C, Hou J, Wang L, Zhang H, Zhang S, Chen X, Zhang X (2015) A yield-associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis. Theor Appl Genet 128:131–143. https://doi.org/10.1007/s00122-014-2417-5
Jobson EM, Martin JM, Schneider TM, Giroux MJ (2018) The impact of the Rht-B1b, Rht-D1b, and Rht-8 wheat semi-dwarfing genes on flour milling, baking, and micronutrients. Cereal Chem 95:770–778. https://doi.org/10.1002/cche.10091
Kato K, Miura H, Sawada S (2000) Mapping QTLs controlling grain yield and its components on chromosome 5A of wheat. Theor Appl Genet 101:1114–1121. https://doi.org/10.1007/s001220051587
Korzun V, Roder MS, Ganal MW, Worland AJ, Law CN (1998) Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theor Appl Genet 96:1104–1109. https://doi.org/10.1007/s001220050845
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugenic 12:172–175. https://doi.org/10.1111/j.1469-1809.1943.tb02321.x
Kuchel H, Williams KJ, Langridge P, Eagles HA, Jefferies SP (2007) Genetic dissection of grain yield in bread wheat. I. QTL analysis. Theor Appl Genet 115:1029–1041. https://doi.org/10.1007/s00122-007-0629-7
Kumar A, Mantovani EE, Seetan R, Soltani A, Echeverry-Solarte M, Jain S, Simsek S, Doehlert D, Alamri MS, Elias EM, Kianian SF, Mergoum M (2016) Dissection of genetic factors underlying wheat kernel shape and size in an elite × nonadapted cross using a high density SNP linkage map. Plant Genome. https://doi.org/10.3835/plantgenome2015.09.0081
Kumari S, Jaiswal V, Mishra VK, Paliwal R, Balyan HS, Gupta PK (2018) QTL mapping for some grain traits in bread wheat (Triticum aestivum L.). Physiol Mol Biol Pla 24:909–920. https://doi.org/10.1007/s12298-018-0552-1
Li N, Li Y (2016) Signaling pathways of seed size control in plants. Curr Opin Plant Biol 33:23–32. https://doi.org/10.1016/j.pbi.2016.05.008
Li Q, Wan JM (2005) SSRHunter: development of a local searching software for SSR sites. Yi Chuan 27:808–810
Li W, Yang B (2017) Translational genomics of grain size regulation in wheat. Theor Appl Genet 130:1765–1771. https://doi.org/10.1007/s00122-017-2953-x
Liu G, Jia L, Lu L, Qin D, Zhang J, Guan P, Ni Z, Yao Y, Sun Q, Peng H (2014) Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor Appl Genet 127:2415–2432. https://doi.org/10.1007/s00122-014-2387-7
Liu S, Hua L, Dong S, Chen H, Zhu X, Jiang J, Zhang F, Li Y, Fang X, Chen F (2015) OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J 84:672–681. https://doi.org/10.1111/tpj.13025
Liu N, Liu J, Li W, Pan Q, Liu J, Yang X, Yan J, Xiao Y (2018) Intraspecific variation of residual heterozygosity and its utility for quantitative genetic studies in maize. BMC Plant Biol 18:66. https://doi.org/10.1186/s12870-018-1287-4
Ma M, Wang Q, Li Z, Cheng H, Li Z, Liu X, Song W, Appels R, Zhao H (2015) Expression of TaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size. Plant J 83:312–325. https://doi.org/10.1111/tpj.12896
Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X (2016) TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol J 14:1269–1280. https://doi.org/10.1111/pbi.12492
Marklund S, Chaudhary R, Marklund L, Sandberg K, Andersson L (2009) Extensive mtDNA diversity in horses revealed by PCR-SSCP analysis. Anim Genet 26:193–196. https://doi.org/10.1111/j.1365-2052.1995.tb03162.x
McCartney C, Somers D, Humphreys D, Lukow O, Ames N, Noll J, Cloutier S, McCallum B (2005) Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × ’AC Domain’. Genome 48:870–883
McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Xia XC (2017) Catalogue of gene symbols for wheat: 2017 supplement
Mo Y, Vanzetti LS, Hale I, Spagnolo EJ, Guidobaldi F, Al-Oboudi J, Odle N, Pearce S, Helguera M, Dubcovsky J (2018) Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor Appl Genet 131:2021–2035. https://doi.org/10.1007/s00122-018-3130-6
Nadolska-Orczyk A, Rajchel IK, Orczyk W, Gasparis S (2017) Major genes determining yield-related traits in wheat and barley. Theor Appl Genet 130:1081–1098. https://doi.org/10.1007/s00122-017-2880-x
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261. https://doi.org/10.1038/22307
Quarrie S, Pekic Quarrie S, Radosevic R, Rancic D, Kaminska A, Barnes JD, Leverington M, Ceoloni C, Dodig D (2006) Dissecting a wheat QTL for yield present in a range of environments: from the QTL to candidate genes. J Exp Bot 57:2627–2637. https://doi.org/10.1093/jxb/erl026
Raihan MS, Liu J, Huang J, Guo H, Pan Q, Yan J (2016) Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 x SK maize population. Theor Appl Genet 129:1465–1477. https://doi.org/10.1007/s00122-016-2717-z
Ramirez-Gonzalez RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, Khedikar Y, Robinson SJ, Cory AT, Florio T, Concia L, Juery C, Schoonbeek H, Steuernagel B, Xiang D, Ridout CJ, Chalhoub B, Mayer KFX, Benhamed M, Latrasse D, Bendahmane A, International Wheat Genome Sequencing C, Wulff BBH, Appels R, Tiwari V, Datla R, Choulet F, Pozniak CJ, Provart NJ, Sharpe AG, Paux E, Spannagl M, Brautigam A, Uauy C (2018) The transcriptional landscape of polyploid wheat. Science 361:662. https://doi.org/10.1126/science.aar6089
Ramya P, Chaubal A, Kulkarni K, Gupta L, Kadoo N, Dhaliwal HS, Chhuneja P, Lagu M, Gupta V (2010) QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J Appl Genet 51:421–429
Roder MS, Huang XQ, Borner A (2008) Fine mapping of the region on wheat chromosome 7D controlling grain weight. Funct Integr Genomics 8:79–86. https://doi.org/10.1007/s10142-007-0053-8
Simmonds J, Scott P, Leverington-Waite M, Turner AS, Brinton J, Korzun V, Snape J, Uauy C (2014) Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat (Triticum aestivum L.). BMC Plant Biol 14:191. https://doi.org/10.1186/s12870-014-0191-9
Simmonds J, Scott P, Brinton J, Mestre TC, Bush M, Del Blanco A, Dubcovsky J, Uauy C (2016) A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor Appl Genet 129:1099–1112. https://doi.org/10.1007/s00122-016-2686-2
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630. https://doi.org/10.1038/ng2014
Su Q, Zhang X, Zhang W, Zhang N, Song L, Liu L, Xue X, Liu G, Liu J, Meng D, Zhi L, Ji J, Zhao X, Yang C, Tong Y, Liu Z, Li J (2018) QTL detection for kernel size and weight in bread wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front Plant Sci 9:1484. https://doi.org/10.3389/fpls.2018.01484
Sukumaran S, Lopes M, Dreisigacker S, Reynolds M (2018) Correction to: genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet 131:999. https://doi.org/10.1007/s00122-018-3066-x
Tian X, Li X, Zhou W, Ren Y, Wang Z, Liu Z, Tang J, Tong H, Fang J, Bu Q (2017a) Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol 175:1337–1349. https://doi.org/10.1104/pp.17.00946
Tian X, Wen W, Xie L, Fu L, Xu D, Fu C, Wang D, Chen X, Xia X, Chen Q, He Z, Cao S (2017b) Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front Plant Sci 8:1379. https://doi.org/10.3389/fpls.2017.01379
Tuinstra MR, Ejeta G, Goldsbrough PB (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95:1005–1011. https://doi.org/10.1007/s001220050654
Van Ooijen W (2006) JoinMap 4.0: Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Van Os H, Stam P, Visser RG, Van Eck HJ (2005) RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet 112:30–40. https://doi.org/10.1007/s00122-005-0097-x
Wang RX, Hai L, Zhang XY, You GX, Yan CS, Xiao SH (2009) QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai x Yu8679. Theor Appl Genet 118:313–325. https://doi.org/10.1007/s00122-008-0901-5
Wang Z, Wu X, Ren Q, Chang X, Li R, Jing R (2010) QTL mapping for developmental behavior of plant height in wheat (Triticum aestivum L.). Euphytica 174:447–458. https://doi.org/10.1007/s10681-010-0166-3
Wang S, Basten C, Zeng Z (2012a) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh 2010
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012b) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954. https://doi.org/10.1038/ng.2327
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796. https://doi.org/10.1111/pbi.12183
Winfield MO, Allen AM, Burridge AJ, Barker GL, Benbow HR, Wilkinson PA, Coghill J, Waterfall C, Davassi A, Scopes G (2016) High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol J 14:1195–1206
Wu QH, Chen YX, Zhou SH, Fu L, Chen JJ, Xiao Y, Zhang D, Ouyang SH, Zhao XJ, Cui Y (2015) High-density genetic linkage map construction and QTL mapping of grain shape and size in the wheat population Yanda 1817 x Beinong6. PLoS ONE 10:e0118144. https://doi.org/10.1371/journal.pone.0118144
Wurschum T, Langer SM, Longin CF (2015) Genetic control of plant height in European winter wheat cultivars. Theor Appl Genet 128:865–874. https://doi.org/10.1007/s00122-015-2476-2
Wurschum T, Leiser WL, Langer SM, Tucker MR, Longin CFH (2018) Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Theor Appl Genet 131:2071–2084. https://doi.org/10.1007/s00122-018-3133-3
Xiao Y, He S, Yan J, Zhang Y, Zhang Y, Wu Y, Xia X, Tian J, Ji W, He Z (2011) Molecular mapping of quantitative trait loci for kernel morphology traits in a non-1BL. 1RS × 1BL. 1RS wheat cross. Crop Pasture Sci 62:625–638
Xie Q, Mayes S, Sparkes DL (2015) Carpel size, grain filling, and morphology determine individual grain weight in wheat. J Exp Bot 66:6715–6730. https://doi.org/10.1093/jxb/erv378
Yan JB, Tang H, Huang YQ, Zheng YL, Li JS (2006) Quantitative trait loci mapping and epistatic analysis for grain yield and yield components using molecular markers with an elite maize hybrid. Euphytica 149:121–131. https://doi.org/10.1007/s10681-005-9060-9
Yan L, Liang F, Xu H, Zhang X, Zhai H, Sun Q, Ni Z (2017) Identification of QTL for grain size and shape on the D genome of natural and synthetic allohexaploid wheats with near-identical AABB genomes. Front Plant Sci 8:1705. https://doi.org/10.3389/fpls.2017.01705
Yang J, Zhou Y, Wu Q, Chen Y, Zhang P, Zhang Y, Hu W, Wang X, Zhao H, Dong L, Han J, Liu Z, Cao T (2019) Molecular characterization of a novel TaGL3-5A allele and its association with grain length in wheat (Triticum aestivum L.). Theor Appl Genet 132:1799–1814. https://doi.org/10.1007/s00122-019-03316-1
Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Neumann F, Eichhorn A, Polley A, Jaenecke C, Ganal MW, Roder MS (2015) Analysis of main effect QTL for thousand grain weight in European winter wheat (Triticum aestivum L.) by genome-wide association mapping. Front Plant Sci 6:644. https://doi.org/10.3389/fpls.2015.00644
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhai HJ, Feng ZY, Li J, Liu XY, Xiao SH, Ni ZF, Sun QX (2016) QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01617
Zhai H, Feng Z, Du X, Song Y, Liu X, Qi Z, Song L, Li J, Li L, Peng H, Hu Z, Yao Y, Xin M, Xiao S, Sun Q, Ni Z (2018) A novel allele of TaGW2-A1 is located in a finely mapped QTL that increases grain weight but decreases grain number in wheat (Triticum aestivum L.). Theor Appl Genet 131:539–553. https://doi.org/10.1007/s00122-017-3017-y
Zhang L, Zhao YL, Gao LF, Zhao GY, Zhou RH, Zhang BS, Jia JZ (2012) TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol 195:574–584. https://doi.org/10.1111/j.1469-8137.2012.04194.x
Zhang JJ, Dell B, Biddulph B, Drake-Brockman F, Walker E, Khan N, Wong DB, Hayden M, Appels R (2013) Wild-type alleles of Rht-B1 and Rht-D1 as independent determinants of thousand-grain weight and kernel number per spike in wheat. Mol Breeding 32:771–783. https://doi.org/10.1007/s11032-013-9905-1
Zhang C, Gao L, Sun J, Jia J, Ren Z (2014a) Haplotype variation of Green Revolution gene Rht-D1 during wheat domestication and improvement. J Integr Plant Biol 56:774–780. https://doi.org/10.1111/jipb.12197
Zhang YJ, Liu JD, Xia XC, He ZH (2014b) TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol Breeding 34:1097–1107. https://doi.org/10.1007/s11032-014-0102-7
Zheng J, Liu H, Wang Y, Wang L, Chang X, Jing R, Hao C, Zhang X (2014) TEF-7A, a transcript elongation factor gene, influences yield-related traits in bread wheat (Triticum aestivum L.). J Exp Bot 65:5351–5365. https://doi.org/10.1093/jxb/eru306
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2016YFD0100801 and 2017YFD0101000).
Author information
Authors and Affiliations
Contributions
ZN conceived the project; XC developed the RIL population; ZC and JL constructed the linkage map. ZC, XC, ZW, JL and RB collected data of RIL population under six environments; ZC and ZW developed markers of the QTL region of interest; ZC developed the near isogenic lines; ZC and LC performed phenotyping of the near isogenic lines; AZ, MX, WG, ZH, HP, YY and QS assisted in revising the manuscript; ZC and ZN analyzed experimental results and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Susanne Dreisigacker.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Histograms of the HS2/4332 recombinant inbred population for TGW, GL, GW and PHT based on the mean values. The Y-axis represents the density (the radio of frequency to group distance) of each trait. P < 0.05 indicates a significant departure from the normal distribution (Shapiro–Wilk test) (TIFF 247 kb)
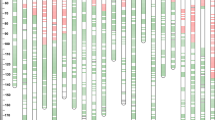
Fig. S2
Illustration of 21 wheat chromosomes in the HS2/4332 RIL population. A centimorgan (cM) scale is shown on the left. The black lines on individual chromosomes represent SNP or SSR markers, the marker names are not shown (TIFF 347 kb)
Fig. S3
Comparison of TGW and GW between lines with HS2 allele and lines with 4332 allele in the RIL population of six environments and BLUP, when excluding the influence of the major QTL QTgw.cau-3D1. Significant differences are indicated by * (P < 0.05), ** (P < 0.01), **** (P < 0.0001) (Student’s t test) (TIFF 539 kb)
Rights and permissions
About this article
Cite this article
Chen, Z., Cheng, X., Chai, L. et al. Dissection of genetic factors underlying grain size and fine mapping of QTgw.cau-7D in common wheat (Triticum aestivum L.). Theor Appl Genet 133, 149–162 (2020). https://doi.org/10.1007/s00122-019-03447-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03447-5