Abstract
Low-temperature germination (LTG) is an important agronomic trait for direct seeding of rice in temperate regions of East Asia. To dissect the genetic control of LTG, we constructed a recombinant inbred line (RIL) population derived from a cross of japonica variety USSR5 and indica variety N22. Three putative QTL involved in LTG were detected and named qLTG-7, qLTG-9 and qLTG-12. They explained 9.5, 12.12 and 7.08 % of the phenotypic variation, respectively, and the alleles from USSR5 enhanced LTG. A set of advanced backcross lines selected for the presence of qLTG-9 (with the biggest contribution of the three QTL), by both linked markers and phenotype, was used to validate qLTG-9 in different generations, years and locations. A near-isogenic line in USSR5 background with a qLTG-9 insertion from N22 had retarded germination under low-temperature conditions. Finally, qLTG-9 was fine mapped between markers L9-25D and ID-1, to a 72.3-kb region in chromosome 9, which in the Nipponbare genome contains five predicted genes. This result provides a springboard for map-based cloning of qLTG-9 and is helpful in understanding the mechanism of seed germination under low-temperature conditions.







Similar content being viewed by others
Abbreviations
- AB:
-
Advanced backcross
- NIL:
-
Near-isogenic line
- MAS:
-
Marker-assisted selection
- MAB:
-
Marker-assisted backcrossing
- PS:
-
Phenotypic selection
- RIL:
-
Recombinant inbred line
- LTG:
-
Low-temperature germination
- QTL:
-
Quantitative trait loci/locus
- qLTG-*:
-
Quantitative trait allele for low-temperature germination
- Chr:
-
Chromosome
- SSR:
-
Simple sequence repeat
- PCR:
-
Polymerase chain reaction
References
Agirregoitia N, Casis L, Gil J, Ruiz F, Irazusta J (2007) Ontogeny of prolyl endopeptidase and pyroglutamyl peptidase I in rat tissues. Regul Pept 139:52–58
Argyris J, Truco MJ, Ochoa O, Knapp SJ, Still DW, Lenssen GM, Schut JW, Michelmore RW, Bradford KJ (2005) Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor Appl Genet 111:1365–1376
Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006) Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103:17042–17047
Brunings AM, Datnoff LE, Ma JF, Mitani N, Nagamura Y, Rathinasabapathi B, Kirst M (2009) Differential gene expression of rice in response to silicon and rice blast fungus Magnaporthe oryzae. Ann Appl Biol 155:161–170
Chen X, Temnykh S, Xu Y, Cho Y, McCouch S (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor Appl Genet 95:553–567
Chen F, Nonogaki H, Bradford KJ (2002) A gibberellin-regulated xyloglucan endotransglycosylase gene is expressed in the endosperm cap during tomato seed germination. J Exp Bot 53:215–223
Chen L, Lou QJ, Sun ZX, Xing YZ, Yu XQ, Luo LJ (2006) QTL mapping of low temperature on germination rate of rice. Rice Sci 13:93–98
Cho SK, Kim JE, Park JA, Eom TJ, Kim WT (2006) Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett 580:3136–3144
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
De Gandarias JM, Irazusta J, Fernandez D, Varona A, Casis L (1994) Developmental changes of pyroglutamate-peptidase I activity in several regions of the female and the male rat brain. Int J Neurosci 77:53–60
De Gandarias JM, Irazusta J, Gil J, Varona A, Ortega F, Casis L (2000) Subcellular ontogeny of brain pyroglutamyl peptidase I. Peptides 21:509–517
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Reptr 1:19–21
Dias PMB, Brunel-Muguet S, Durr C, Huguet T, Demilly D, Wagner MH, Teulat-Mera B (2011) QTL analysis of seed germination and pre-emergence growth at extreme temperatures in Medicago truncatula. Theor Appl Genet 122:429–444
Fujino K, Matsuda Y (2010) Genome-wide analysis of genes targeted by qLTG3-1 controlling low-temperature germinability in rice. Plant Mol Biol 72:137–152
Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y, Takeuchi Y, Ando T, Lin SY, Sugimoto M (2004) Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor Appl Genet 108:794–799
Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Sugimoto M (2008) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA 105:12623–12628
Fuse Y, Polk DH, Lam RW, Reviczky AL, Fisher DA (1990) Distribution and ontogeny of thyrotropin-releasing hormone degrading enzymes in rats. Am J Physiol 259(6 Pt 1):E787–E791
Hayashi E, Aoyama N, Still DW (2008) Quantitative trait loci associated with lettuce seed germination under different temperature and light environments. Genome 51:928–947
Hou MY, Wang CM, Jiang L, Wan JM, Yasui H, Yoshimura A (2004) Inheritance and QTL mapping of low temperature germinability in rice (Oryza sativa L.). Acta Genet Sin 31:701–706
Hu GY, Zhao JM, Zhou B, Zuo QM, Gai JY, Yu DY, Xing H (2008) Inheritance and molecular marker of chilling tolerance of soybean in early stage. Soybean Sci 27:905–910
Iwata N, Fujino K (2010) Genetic effects of major QTLs controlling low-temperature germinability in different genetic backgrounds in rice (Oryza sativa L.). Genome 53:763–768
Ji SL, Jiang L, Wang YH, Liu SJ, Liu X, Zhai HQ, Yoshimura A, Wan JM (2008) QTL and epistasis for low temperature germinability in rice. Acta Agron Sin 34:551–556
Ji SL, Jiang L, Wang YH, Zhang WW, Liu X, Liu SJ, Chen LM, Zhai HQ, Wan JM (2009) Quantitative trait loci mapping and stability for low temperature germination ability of rice. Plant Breed 128:387–392
Jiang L, Liu SJ, Hou MY, Tang JY, Chen LM, Zhai HQ, Wan JM (2006) Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.). Field Crops Res 98:68–75
Jiang HW, Li CD, Liu CY, Zhang WB, Qiu PC, Li WF, Gao YL, Hu GH, Chen QS (2009) Genotype analysis and QTL mapping for tolerance to low temperature in germination by introgression lines in soybean. Acta Agron Sin 35:1268–1273
Jin J, Huang W, Gao JP, Yang J, Shi M, Zhu MZ, Luo D, Lin HX (2008) Genetic control of rice plant architecture under domestication. Nat Genet 40:1365–1369
Lin SY, Sasaki T, Yano M (1998) Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet 96:997–1003
Lin HX, Ashikari M, Yamanouchi U, Sasaki T, Yano M (2002) Identification and characterization of a quantitative trait locus, Hd9, controlling heading date in rice. Breed Sci 52:35–41
Lu B, Xie K, Yang C, Wang S, Liu X, Zhang L, Jiang L, Wan J (2011) Mapping two major effect grain dormancy QTL in rice. Mol Breed 28:453–462
McCouch S, Cho Y, Sugimoto M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14
McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu BY, Maghirang R, Li ZK, Xing YZ, Zhang QF, Kono I, Sugimoto M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Ren ZH, Zheng ZM, Chinnusamy V, Zhu JH, Cui XP, Iida K, Zhu JK (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 107:10330
Sanguinetti C, Dias NE, Simpson A (1994) Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17:914
Sasaki T, Honma H (1974) Studies on breeding for the germinability at low temperature of rice varieties adapted to direct sowing cultivation in flooded paddy field in cool region. Bull Hokkaido Pref Agric Exp Station 24:1–90
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Sugimoto M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40:1023–1028
Sugimoto K, Takeuchi Y, Ebana K, Miyao A, Hirochika H, Hara N, Ishiyama K, Kobayashi M, Ban Y, Hattori T, Sugimoto M (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA 107:5792–5797
Sun Q, Wang JH, Sun BQ (2007) Advances on seed vigor physiological and genetic mechanisms. Agric Sci China 6:1060–1066
Tanksley SD, Young ND, Paterson AH, Bonierbale MW (1989) RFLP mapping in plant breeding: new tools for an old science. Biotechnology 7:257–264
Tanksley S, Grandillo S, Fulton T, Zamir D, Eshed Y, Petiard V, Lopez J, Beck-Bunn T (1996) Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor Appl Genet 92:213–224
Teng S, Zeng D, Qian Q, Kunihifo Y, Huang D, Zhu L (2001) QTL analysis of rice low temperature germinability. Chin Sci Bull 46:1800–1803
Thomson MJ, Ismail AM, McCouch SR, Mackill DJ (2010) Marker assisted breeding. In: Pareen A, Sopory SK, Bohner HJ, Govindjee (eds) Abiotic stress adaptation in plants: physiological, molecular and genomic foundation. Springer SBM, Dordrecht, pp 451–469
Wan JM, Jiang L, Tang JY, Wang CM, Hou MY, Jing W, Zhang L (2006) Genetic dissection of the seed dormancy trait in cultivated rice (Oryza sativa L.). Plant Sci 170:786–792
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
Wu WX, Zheng XM, Lu GW, Zhong ZZ, Gao H, Chen LP, Wu CY, Wang HJ, Wang Q, Zhou KN, WangJL WuFQ, Zhang X, Guo XP, Cheng ZJ, Lei CL, Lin QB, Jiang L, Wang HY, Ge S, Wan JM (2013) Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA 110:2775–2780
Xie K, Jiang L, Lu BY, Yang CY, Li LF, Liu X, Zhang L, Zhao ZG, Wan J (2011) Identification of QTLs for seed dormancy in rice (Oryza sativa L.). Plant Breed 130:328–332
Yamamoto T, Lin HX, Sasaki T, Yano M (2000) Identification of heading date quantitative trait locus Hd6, and characterization of its epistatic interaction with Hd2 in rice using advanced backcross progeny. Genetics 154:885–891
Yano M, Harushima Y, Nagamura Y, Kurata N, Minobe Y, Sasaki T (1997) Identification of quantitative trait loci controlling heading date in rice using a high density linkage map. Theor Appl Genet 95:1025–1032
Yokoyama R, Uwagaki Y, Sasaki H, Harada T, Hiwatashi Y, Hasebe M, Nishitani K (2010) Biological implications of the occurrence of 32 members of the XTH (xyloglucan endotransglucosylase/hydrolase) family of proteins in the bryophyte Physcomitrella patens. Plant J 64:645–656
Zhang WB, Qiu PC, Jiang HW, Liu CY, Xin DW, Li CD, Hu GH, Chen QS (2012) Dissection of genetic overlap of drought and low-temperature tolerance QTLs at the germination stage using backcross introgression lines in soybean. Mol Biol Rep 39:6087–6094
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (No. 30671246), 863 Program (projects 2012AA101101, 2011AA10A101) of China, Jiangsu Cultivar Development Program (projects BE2012303, BK2010016), Jiangsu Independence Innovation Project (CX(12)1003) and PAPD program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wissuwa.
Electronic supplementary material
122_2013_2137_MOESM1_ESM.doc
Fig. S1. Chromosome locations of low temperature germination quantitative trait loci in RIL at the fourth and fifth days of germination (DOC 126 kb)
122_2013_2137_MOESM2_ESM.doc
Fig. S2. Genetic backgrounds of W907 (BC4F2) (A), Y2469 (BC4F3) (B), Y2288 (BC5F2) (C). Black square represent fragments from N22. Hollow square are QTL detected in RIL (DOC 26 kb)
122_2013_2137_MOESM3_ESM.doc
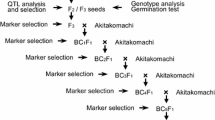
Fig. S3. Breeding scheme employed to develop the advanced backcross lines and mapping populations. Numbers in brackets represent the size of each population. PS: Phenotypic selection. MAS + PS: Marker assistant selection plus phenotypic selection (DOC 717 kb)
Rights and permissions
About this article
Cite this article
Li, L., Liu, X., Xie, K. et al. qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor Appl Genet 126, 2313–2322 (2013). https://doi.org/10.1007/s00122-013-2137-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2137-2