Abstract
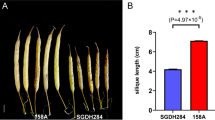
Silique length (SL) and seed weight (SW) are two important yield-related traits controlled by quantitative trait loci (QTL) in oilseed rape (Brassica napus L.). The genetic bases underlying these two traits are largely unknown at present. In this study, we conducted QTL analyses for SL and SW using 186 recombinant inbred lines (RILs) derived from a cross between S1, an EMS mutant with extremely long siliques and large seeds, and S2, an inbred line with regular silique length and seed size. RILs were grown in Wuhan in the 2008/09 (SS09) and 2009/10 (SS10) growing seasons, and mean SL and SW for each line were investigated. Ten non-redundant QTL were identified for SL. Of these, a major QTL, cqSLA9, consistently explained as much as 53.4% of SL variation across environments. The others are minor QTL and individually explained less than 10% of the SL variation. Nine non-redundant QTL were identified for SW. Of which, one major QTL, cqSWA9, explained as much as 28.2% of the total SW variation in the SS09 and SS10 environments. In addition, three additive by additive interactions with small effects were detected for SL, and no interactions were detected for SW. Interestingly, the two major QTL, cqSLA9 for SL and cqSWA9 for SW colocalized in the same chromosomal region and were integrated into a unique QTL, uqA9. The S1 allele at this locus increases both SL and SW, suggesting that uqA9 has pleiotropic effects on both SL and SW. The existence and effect of uqA9 was confirmed in genetically different RILs derived from the cross between S1 and No2127, a resynthesized DH line having regular silique length and seed size. Individuals in one residual heterozygous line for cqSLA9 showed significant difference in silique length. The results in this study revealed that silique length in the S1 mutant is mainly controlled by the cqSLA9 locus, which will be suitable for fine mapping and marker-assisted selection in rapeseed breeding for high yield.



Similar content being viewed by others
References
Allen E, Morgan D, Ridgman W (1971) A physiological analysis of the growth of oilseed rape. J Agric Sci 77:339–341
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20:2324–2326
Aytac Z, Kinaci G (2009) Genetic variability and association studies of some quantitative characters in winter rapeseed (Brassica napus L.). Afr J Biotechnol 8:3547–3554
Basunanda P, Radoev M, Ecke W, Friedt W, Becker H, Snowdon R (2010) Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). Theor Appl Genet 120:271–281
Bennett EJ, Roberts JA, Wagstaff C (2011) The role of the pod in seed development: strategies for manipulating yield. New Phytol 190:838–853
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664
Brar G, Thies W (1977) Contribution of leaves, stem, siliques and seeds to dry matter accumulation in ripening seeds of rapeseed, Brassica napus L. Z Pflanzenphysiol 82:1–13
Butruille DV, Guries RP, Osborn TC (1999) Linkage analysis of molecular markers and quantitative trait loci in populations of inbred backcross lines of Brassica napus L. Genetics 153:949–964
Chay P, Thurling N (1989) Identification of genes controlling pod length in spring rapeseed, Brassica napus L., and their utilization for yield improvement. Plant Breed 103:54–62
Chen B, Heneen W, J nsson R (1988) Resynthesis of Brassies napus L. through interspecific hybridization between B. alboglabra Bailey and B. campestris L. with special emphasis on seed colour. Plant Breed 101:52–59
Chen W, Zhang Y, Liu X, Chen B, Tu J, Fu T (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor Appl Genet 115:849–858
Cheng X, Xu J, Xia S, Gu J, Yang Y, Fu J, Qian X, Zhang S, Wu J, Liu K (2009) Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor Appl Genet 118:1121–1131
Choi SR, Teakle GR, Plaha P, Kim JH, Allender CJ, Beynon E, Piao ZY, Soengas P, Han TH, King GJ, Barker GC, Hand P, Lydiate DJ, Batley J, Edwards D, Koo DH, Bang JW, Park BS, Lim YP (2007) The reference genetic linkage map for the multinational Brassica rapa genome sequencing project. Theor Appl Genet 115:777–792
Clarke J, Simpson G (1978) Influence of irrigation and seeding rates on yield and yield components of Brassica napus cv. Tower. Can J Plant Sci 58:731–737
Diepenbrock W (2000) Yield analysis of winter oilseed rape (Brassica napus L.): a review. Field Crops Res 67:35–49
Fan C, Cai G, Qin J, Li Q, Yang M, Wu J, Fu T, Liu K, Zhou Y (2010) Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor Appl Genet 121:1289–1301
Goffinet B, Gerber S (2000) Quantitative trait loci: a meta-analysis. Genetics 155:463–473
Hasan M, Friedt W, Pons-Kuhnemann J, Freitag NM, Link K, Snowdon RJ (2008) Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus). Theor Appl Genet 116:1035–1049
Kashiani P, Saleh G (2010) Estimation of genetic correlations on sweet corn inbred lines using SAS mixed model. Am J Agr Biol Sci 5:309–314
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugenic 12:172–175
Kresovich S, Szewc-McFadden A, Bliek S, McFerson J (1995) Abundance and characterization of simple-sequence repeats (SSRs) isolated from a size-fractionated genomic library of Brassica napus L. (rapeseed). Theor Appl Genet 91:206–211
Lagercrantz U, Ellegren H, Andersson L (1993) The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res 21:1111
Latta RG, Gardner KM (2007) Shared quantitative trait loci underlying the genetic correlation between continuous traits. Mol Ecol 16:4195–4209
Lebowitz R (1989) Image analysis measurements and repeatability estimates of siliqua morphological traits in Brassica campestris L. Euphytica 43:113–116
Li H, Chen X, Yang Y, Xu J, Gu J, Fu J, Qian X, Zhang S, Wu J, Liu K (2011) Development and genetic mapping of microsatellite markers from whole genome shotgun sequences in Brassica oleracea. Mol Breed 28:585–596
Lowe AJ, Jones AE, Raybould AF, Trick M, Moule CL, Edwards KJ (2002) Transferability and genome specificity of a new set of microsatellite primers among Brassica species of the U triangle. Mol Ecol Notes 2:7–11
Lowe AJ, Moule C, Trick M, Edwards KJ (2004) Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor Appl Genet 108:1103–1112
Miura K, Ashikari M, Matsuoka M (2011) The role of QTLs in the breeding of high-yielding rice. Trends Plant Sci 16:319–326
Özer H, Oral E, Dogru Ü (1999) Relationships between yield and yield components on currently improved spring rapeseed cultivars. Turk J Agric For 23:603–608
Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Piquemal J, Cinquin E, Couton F, Rondeau C, Seignoret E, Doucet I, Perret D, Villeger MJ, Vincourt P, Blanchard P (2005) Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor Appl Genet 111:1514–1523
Quijada PA, Udall JA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet 113:549–561
Radoev M, Becker HC, Ecke W (2008) Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179:1547–1558
Samizadeh H, Yazdi-Samadi B, Ghannadha MR, Malbobi MA, Taleei AR, Stringam GR (2007) Study of pod length trait in doubled haploid brassica napus population by molecular markers. J Agric Sci Technol 9:129–136
SAS Institute (2000) SAS/STAT user’s guide, version 8. SAS Institute, Cary
Schiltz S, Munier-Jolain N, Jeudy C, Burstin J, Salon C (2005) Dynamics of exogenous nitrogen partitioning and nitrogen remobilization from vegetative organs in pea revealed by 15N in vivo labeling throughout seed filling. Plant Physiol 137:1463–1473
Shi S, Wu J, Liu H (1995) Dwarf stem and long pod mutants induced from microspore embryo cultures of Brassica napus with EMS. Acta Agr Nucl Sin 9:252–253
Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182:851–861
Singh J, Yadav M, Sheikh I (1996) Genetical studies for yield and oil content in Brassica juncea (L.) Czern and Coss. Indian J Genet 56:299–304
Snowdon R, Lühs W, Friedt W (2007) Oilseed rape. Springer, Berlin, pp 55–114
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S (2006) Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173:309–319
Suwabe K, Morgan C, Bancroft I (2008) Integration of Brassica A genome genetic linkage map between Brassica napus and B. rapa. Genome 51:169–176
Szewc-McFadden A, Kresovich S, Bliek S, Mitchell S, McFerson J (1996) Identification of polymorphic, conserved simple sequence repeats (SSRs) in cultivated Brassica species. Theor Appl Genet 93:534–538
Takeda S, Matsuoka M (2008) Genetic approaches to crop improvement: responding to environmental and population changes. Nat Rev Genet 9:444–457
Udall JA, Quijada PA, Lambert B, Osborn TC (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor Appl Genet 113:597–609
Uzunova M, Ecke W (1999) Abundance, polymorphism and genetic mapping of microsatellites in oilseed rape (Brassica napus L.). Plant Breed 118:323–326
Van Ooijen J, Voorrips R (2001) JoinMap(R) 3.0. Software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Wang F, Wang X, Chen X, Xiao Y, Li H, Zhang S, Xu J, Fu J, Huang L, Liu C, Wu J, Liu K (2011) Abundance, marker development and genetic mapping of microsatellites from unigenes in Brassica napus. Mol Breed. doi:10.1007/s11032-011-9658-7
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Xu J, Qian X, Wang X, Li R, Cheng X, Yang Y, Fu J, Zhang S, King GJ, Wu J, Liu K (2010) Construction of an integrated genetic linkage map for the A genome of Brassica napus using SSR markers derived from sequenced BACs in B. rapa. BMC Genomics 11:594
Yamanaka N, Watanabe S, Toda K, Hayashi M, Fuchigami H, Takahashi R, Harada K (2005) Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor Appl Genet 110:634–639
Yang J, Hu C, Hu H, Yu R, Xia Z, Ye X, Zhu J (2008) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723
Yin Y, Wang H, Liao X (2009) Analysis and strategy for 2009 rapeseed industry development in China. Chin J Oil Crop Sci 31:259–262
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang L, Yang G, Liu P, Hong D, Li S, He Q (2011) Genetic and correlation analysis of silique-traits in Brassica napus L. by quantitative trait locus mapping. Theor Appl Genet 122:21–31
Acknowledgments
The research was supported by the National Natural Science Foundation of China (No. 31071452) and the Doctoral Fund of Ministry of Education of China (No. 20100146110019).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Snape.
Rights and permissions
About this article
Cite this article
Yang, P., Shu, C., Chen, L. et al. Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.). Theor Appl Genet 125, 285–296 (2012). https://doi.org/10.1007/s00122-012-1833-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1833-7