Abstract
Diabetic nephropathy (DN), an important complication of diabetic microvascular disease, is one of the leading causes of end-stage renal disease (ESRD), which brings heavy burdens to the whole society. Podocytes are terminally differentiated glomerular cells, which act as a pivotal component of glomerular filtration barrier. When podocytes are injured, glomerular filtration barrier is damaged, and proteinuria would occur. Dysfunction of podocytes contributes to DN. And degrees of podocyte injury influence prognosis of DN. Growing evidences have shown that epigenetics does a lot in the evolvement of podocyte injury. Epigenetics includes DNA methylation, histone modification, and non-coding RNA. Among them, histone modification plays an indelible role. Histone modification includes histone methylation, histone acetylation, and other modifications such as histone phosphorylation, histone ubiquitination, histone ADP-ribosylation, histone crotonylation, and histone β-hydroxybutyrylation. It can affect chromatin structure and regulate gene transcription to exert its function. This review is to summarize documents about pathogenesis of podocyte injury, most importantly, histone modification of podocyte injury in DN recently to provide new ideas for further molecular research, diagnosis, and treatment.


Similar content being viewed by others
Availability of data and materials
Not applicable.
References
Qi C, Mao X, Zhang Z, Wu H (2017) Classification and differential diagnosis of diabetic nephropathy. J Diabetes Res 2017:8637138
Nagata M (2016) Podocyte injury and its consequences. Kidney Int 89(6):1221–1230
White KE, Bilous RW (2004) Structural alterations to the podocyte are related to proteinuria in type 2 diabetic patients. Nephrol Dial Transplant 19(6):1437–1440
Lan J, Lepikhov K, Giehr P, Walter J (2017) Histone and DNA methylation control by H3 serine 10/threonine 11 phosphorylation in the mouse zygote. Epigenetics Chromatin 10:5
Nathan DM (2014) The diabetes control and complications trial/epidemiology of diabetes interventions and complications study at 30 years: overview. Diabetes Care 37(1):9–16
Kato M, Natarajan R (2019) Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol 15(6):327–345
Epidemiology of Diabetes Interventions and Complications (EDIC) (1999) Design, implementation, and preliminary results of a long-term follow-up of the Diabetes Control and Complications Trial cohort. Diabetes Care 22(1): 99–111
Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A (2009) An operational definition of epigenetics. Genes Dev 23(7):781–783
Lu Z, Liu N, Wang F (2017) Epigenetic regulations in diabetic nephropathy. J Diabetes Res 2017:7805058
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403(6765):41–45
Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98(3):285–294
Voigt P, Reinberg D (2011) Histone tails: ideal motifs for probing epigenetics through chemical biology approaches. ChemBioChem 12(2):236–252
Kimura H (2013) Histone modifications for human epigenome analysis. J Hum Genet 58(7):439–445
Jin J, Gong J, Zhao L, Zhang H, He Q, Jiang X (2019) Inhibition of high mobility group box 1 (HMGB1) attenuates podocyte apoptosis and epithelial-mesenchymal transition by regulating autophagy flux. J Diabetes 11(10):826–836
Lu CC, Wang GH, Lu J, Chen PP, Zhang Y, Hu ZB et al (2019) Role of podocyte injury in glomerulosclerosis. Adv Exp Med Biol 1165:195–232
Liapis H, Romagnani P, Anders HJ (2013) New insights into the pathology of podocyte loss: mitotic catastrophe. Am J Pathol 183(5):1364–1374
Kopp JB, Anders HJ, Susztak K, Podesta MA, Remuzzi G, Hildebrandt F et al (2020) Podocytopathies. Nat Rev Dis Primers 6(1):68
Al-Malki AL (2014) Assessment of urinary osteopontin in association with podocyte for early predication of nephropathy in diabetic patients. Dis Markers 2014:493736
Dai H, Liu Q, Liu B (2017) Research progress on mechanism of podocyte depletion in diabetic nephropathy. J Diabetes Res 2017:2615286
Schiffer M, Bitzer M, Roberts IS, Kopp JB, Ten DP, Mundel P et al (2001) Apoptosis in podocytes induced by TGF-beta and Smad7. J Clin Invest 108(6):807–816
Maquigussa E, Paterno JC, de Oliveira PG, Da SPM, Varela VA, Da SNA et al (2018) Klotho and PPAR gamma activation mediate the renoprotective effect of losartan in the 5/6 nephrectomy model. Front Physiol 9:1033
Long YC, Zierath JR (2006) AMP-activated protein kinase signaling in metabolic regulation. J Clin Invest 116(7):1776–1783
Eid AA, Ford BM, Block K, Kasinath BS, Gorin Y, Ghosh-Choudhury G et al (2010) AMP-activated protein kinase (AMPK) negatively regulates Nox4-dependent activation of p53 and epithelial cell apoptosis in diabetes. J Biol Chem 285(48):37503–37512
Susztak K, Raff AC, Schiffer M, Bottinger EP (2006) Glucose-induced reactive oxygen species cause apoptosis of podocytes and podocyte depletion at the onset of diabetic nephropathy. Diabetes 55(1):225–233
Cui FQ, Wang YF, Gao YB, Meng Y, Cai Z, Shen C et al (2019) Effects of BSF on podocyte apoptosis via regulating the ROS-mediated PI3K/AKT pathway in DN. J Diabetes Res 2019:9512406
Chen X, Liu W, Xiao J, Zhang Y, Chen Y, Luo C et al (2020) FOXO3a accumulation and activation accelerate oxidative stress-induced podocyte injury. Faseb J 34(10):13300–13316
Gui D, Guo Y, Wang F, Liu W, Chen J, Chen Y et al (2012) Astragaloside IV, a novel antioxidant, prevents glucose-induced podocyte apoptosis in vitro and in vivo. PLoS ONE 7(6):e39824
Araujo M, Wilcox CS (2014) Oxidative stress in hypertension: role of the kidney. Antioxid Redox Signal 20(1):74–101
Chung SS, Ho EC, Lam KS, Chung SK (2003) Contribution of polyol pathway to diabetes-induced oxidative stress. J Am Soc Nephrol 14(8 Suppl 3):S233–S236
Forbes JM, Cooper ME, Oldfield MD, Thomas MC (2003) Role of advanced glycation end products in diabetic nephropathy. J Am Soc Nephrol 14(8 Suppl 3):S254–S258
Yamagishi S, Matsui T (2010) Advanced glycation end products, oxidative stress and diabetic nephropathy. Oxid Med Cell Longev 3(2):101–108
Ha H, Lee HB (2005) Reactive oxygen species amplify glucose signalling in renal cells cultured under high glucose and in diabetic kidney. Nephrology (Carlton) 10(Suppl):S7–S10
Glick D, Barth S, Macleod KF (2010) Autophagy: cellular and molecular mechanisms. J Pathol 221(1):3–12
Liu N, Shi Y, Zhuang S (2016) Autophagy in chronic kidney diseases. Kidney Dis (Basel) 2(1):37–45
Tagawa A, Yasuda M, Kume S, Yamahara K, Nakazawa J, Chin-Kanasaki M et al (2016) Impaired podocyte autophagy exacerbates proteinuria in diabetic nephropathy. Diabetes 65(3):755–767
Liu N, Xu L, Shi Y, Zhuang S (2017) Podocyte autophagy: a potential therapeutic target to prevent the progression of diabetic nephropathy. J Diabetes Res 2017:3560238
Hartleben B, Godel M, Meyer-Schwesinger C, Liu S, Ulrich T, Kobler S et al (2010) Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest 120(4):1084–1096
Liu J, Li QX, Wang XJ, Zhang C, Duan YQ, Wang ZY et al (2016) beta-Arrestins promote podocyte injury by inhibition of autophagy in diabetic nephropathy. Cell Death Dis 7:e2183
Inoki K, Mori H, Wang J, Suzuki T, Hong S, Yoshida S et al (2011) mTORC1 activation in podocytes is a critical step in the development of diabetic nephropathy in mice. J Clin Invest 121(6):2181–2196
Vollenbroker B, George B, Wolfgart M, Saleem MA, Pavenstadt H, Weide T (2009) mTOR regulates expression of slit diaphragm proteins and cytoskeleton structure in podocytes. Am J Physiol Renal Physiol 296(2):F418–F426
Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A (2005) TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell 16(4):1987–2002
Ha TS (2013) Roles of adaptor proteins in podocyte biology. World J Nephrol 2(1):1–10
Ying Q, Wu G (2017) Molecular mechanisms involved in podocyte EMT and concomitant diabetic kidney diseases: an update. Ren Fail 39(1):474–483
Xing L, Liu Q, Fu S, Li S, Yang L, Liu S et al (2015) PTEN inhibits high glucose-induced phenotypic transition in podocytes. J Cell Biochem 116(8):1776–1784
Xu H, Wang X, Liu M, He X (2017) Tangzhiqing granules alleviate podocyte epithelial-mesenchymal transition in kidney of diabetic rats. Evid Based Complement Alternat Med 2017:1479136
Willis BC, Borok Z (2007) TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol 293(3):L525–L534
Chiang YT, Ip W, Jin T (2012) The role of the Wnt signaling pathway in incretin hormone production and function. Front Physiol 3:273
Dai C, Stolz DB, Kiss LP, Monga SP, Holzman LB, Liu Y (2009) Wnt/beta-catenin signaling promotes podocyte dysfunction and albuminuria. J Am Soc Nephrol 20(9):1997–2008
Ichimura K, Kurihara H, Sakai T (2003) Actin filament organization of foot processes in rat podocytes. J Histochem Cytochem 51(12):1589–1600
Perico L, Conti S, Benigni A, Remuzzi G (2016) Podocyte-actin dynamics in health and disease. Nat Rev Nephrol 12(11):692–710
Mathieson PW (2012) The podocyte cytoskeleton in health and in disease. Clin Kidney J 5(6):498–501
Wieder N, Greka A (2016) Calcium, TRPC channels, and regulation of the actin cytoskeleton in podocytes: towards a future of targeted therapies. Pediatr Nephrol 31(7):1047–1054
Reiser J, Polu KR, Moller CC, Kenlan P, Altintas MM, Wei C et al (2005) TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat Genet 37(7):739–744
Tian D, Jacobo SM, Billing D, Rozkalne A, Gage SD, Anagnostou T et al (2010) Antagonistic regulation of actin dynamics and cell motility by TRPC5 and TRPC6 channels. Sci Signal 3(145):a77
Wang Q, Tian X, Wang Y, Wang Y, Li J, Zhao T et al (2020) Role of transient receptor potential canonical channel 6 (TRPC6) in diabetic kidney disease by regulating podocyte actin cytoskeleton rearrangement. J Diabetes Res 2020:6897390
Sonneveld R, van der Vlag J, Baltissen MP, Verkaart SA, Wetzels JF, Berden JH et al (2014) Glucose specifically regulates TRPC6 expression in the podocyte in an AngII-dependent manner. Am J Pathol 184(6):1715–1726
Yang H, Zhao B, Liao C, Zhang R, Meng K, Xu J et al (2013) High glucose-induced apoptosis in cultured podocytes involves TRPC6-dependent calcium entry via the RhoA/ROCK pathway. Biochem Biophys Res Commun 434(2):394–400
Farmer LK, Rollason R, Whitcomb DJ, Ni L, Goodliff A, Lay AC et al (2019) TRPC6 binds to and activates calpain, independent of its channel activity, and regulates podocyte cytoskeleton, cell adhesion, and motility. J Am Soc Nephrol 30(10):1910–1924
Dryer SE, Roshanravan H, Kim EY (2019) TRPC channels: regulation, dysregulation and contributions to chronic kidney disease. Biochim Biophys Acta Mol Basis Dis 1865(6):1041–1066
Zhou Y, Castonguay P, Sidhom EH, Clark AR, Dvela-Levitt M, Kim S et al (2017) A small-molecule inhibitor of TRPC5 ion channels suppresses progressive kidney disease in animal models. Science 358(6368):1332–1336
Wang X, Dande RR, Yu H, Samelko B, Miller RE, Altintas MM et al (2018) TRPC5 does not cause or aggravate glomerular disease. J Am Soc Nephrol 29(2):409–415
Siddiqi FS, Advani A (2013) Endothelial-podocyte crosstalk: the missing link between endothelial dysfunction and albuminuria in diabetes. Diabetes 62(11):3647–3655
Wang YY, Tang LQ, Wei W (2018) Berberine attenuates podocytes injury caused by exosomes derived from high glucose-induced mesangial cells through TGFbeta1-PI3K/AKT pathway. Eur J Pharmacol 824:185–192
Wu X, Gao Y, Xu L, Dang W, Yan H, Zou D et al (2017) Exosomes from high glucose-treated glomerular endothelial cells trigger the epithelial-mesenchymal transition and dysfunction of podocytes. Sci Rep 7(1):9371
Howlett AC, Blume LC, Dalton GD (2010) CB(1) cannabinoid receptors and their associated proteins. Curr Med Chem 17(14):1382–1393
Horvath B, Mukhopadhyay P, Hasko G, Pacher P (2012) The endocannabinoid system and plant-derived cannabinoids in diabetes and diabetic complications. Am J Pathol 180(2):432–442
Gruden G, Barutta F, Kunos G, Pacher P (2016) Role of the endocannabinoid system in diabetes and diabetic complications. Br J Pharmacol 173(7):1116–1127
Barutta F, Mastrocola R, Bellini S, Bruno G, Gruden G (2018) Cannabinoid receptors in diabetic kidney disease. Curr Diab Rep 18(2):9
Barutta F, Corbelli A, Mastrocola R, Gambino R, Di Marzo V, Pinach S et al (2010) Cannabinoid receptor 1 blockade ameliorates albuminuria in experimental diabetic nephropathy. Diabetes 59(4):1046–1054
Jourdan T, Szanda G, Rosenberg AZ, Tam J, Earley BJ, Godlewski G et al (2014) Overactive cannabinoid 1 receptor in podocytes drives type 2 diabetic nephropathy. Proc Natl Acad Sci U S A 111(50):E5420–E5428
Barutta F, Grimaldi S, Franco I, Bellini S, Gambino R, Pinach S et al (2014) Deficiency of cannabinoid receptor of type 2 worsens renal functional and structural abnormalities in streptozotocin-induced diabetic mice. Kidney Int 86(5):979–990
Barutta F, Piscitelli F, Pinach S, Bruno G, Gambino R, Rastaldi MP et al (2011) Protective role of cannabinoid receptor type 2 in a mouse model of diabetic nephropathy. Diabetes 60(9):2386–2396
Barutta F, Grimaldi S, Gambino R, Vemuri K, Makriyannis A, Annaratone L et al (2017) Dual therapy targeting the endocannabinoid system prevents experimental diabetic nephropathy. Nephrol Dial Transplant 32(10):1655–1665
Jorgensen S, Schotta G, Sorensen CS (2013) Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 41(5):2797–2806
Sun GD, Cui WP, Guo QY, Miao LN (2014) Histone lysine methylation in diabetic nephropathy. J Diabetes Res 2014:654148
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119(7):941–953
Nguyen AT, Zhang Y (2011) The diverse functions of Dot1 and H3K79 methylation. Genes Dev 25(13):1345–1358
Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S et al (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449(7163):689–694
Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D et al (2007) Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318(5849):447–450
Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J et al (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449(7163):731–734
Chen H, Huang Y, Zhu X, Liu C, Yuan Y, Su H et al (2019) Histone demethylase UTX is a therapeutic target for diabetic kidney disease. J Physiol 597(6):1643–1660
Liebisch M, Wolf G (2020) AGE-induced suppression of EZH2 mediates injury of podocytes by reducing H3K27me3. Am J Nephrol 51(9):676–692
Majumder S, Thieme K, Batchu SN, Alghamdi TA, Bowskill BB, Kabir MG et al (2018) Shifts in podocyte histone H3K27me3 regulate mouse and human glomerular disease. J Clin Invest 128(1):483–499
Lin CL, Hsu YC, Huang YT, Shih YH, Wang CJ, Chiang WC et al (2019) A KDM6A-KLF10 reinforcing feedback mechanism aggravates diabetic podocyte dysfunction. Embo Mol Med 11(5)
Liu DW, Zhang JH, Liu FX, Wang XT, Pan SK, Jiang DK et al (2019) Silencing of long noncoding RNA PVT1 inhibits podocyte damage and apoptosis in diabetic nephropathy by upregulating FOXA1. Exp Mol Med 51(8):1–15
Hughes AL, Kelley JR, Klose RJ (2020) Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation. Biochim Biophys Acta Gene Regul Mech 1863(8):194567
Lefevre GM, Patel SR, Kim D, Tessarollo L, Dressler GR (2010) Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. Plos Genet 6(10):e1001142
Sayyed SG, Gaikwad AB, Lichtnekert J, Kulkarni O, Eulberg D, Klussmann S et al (2010) Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transplant 25(6):1811–1817
Miao F, Gonzalo IG, Lanting L, Natarajan R (2004) In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem 279(17):18091–18097
Cao A, Li J, Asadi M, Basgen JM, Zhu B, Yi Z et al (2021) DACH1 protects podocytes from experimental diabetic injury and modulates PTIP-H3K4Me3 activity. J Clin Invest 131(10)
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21(3):381–395
Yang XJ, Seto E (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26(37):5310–5318
Shi W, Huang Y, Zhao X, Xie Z, Dong W, Li R et al (2020) Histone deacetylase 4 mediates high glucose-induced podocyte apoptosis via upregulation of calcineurin. Biochem Biophys Res Commun 533(4):1061–1068
Lin CL, Lee PH, Hsu YC, Lei CC, Ko JY, Chuang PC et al (2014) MicroRNA-29a promotion of nephrin acetylation ameliorates hyperglycemia-induced podocyte dysfunction. J Am Soc Nephrol 25(8):1698–1709
Gondaliya P, P DA, Jash K, Tekade RK, Srivastava A, Kalia K (2020) miR-29b attenuates histone deacetylase-4 mediated podocyte dysfunction and renal fibrosis in diabetic nephropathy. J Diabetes Metab Disord 19(1): 13–27
Lundh M, Petersen PS, Isidor MS, Kazoka-Sorensen DN, Plucinska K, Shamsi F et al (2019) Afadin is a scaffold protein repressing insulin action via HDAC6 in adipose tissue. Embo Rep 20(8):e48216
Liang T, Qi C, Lai Y, Xie J, Wang H, Zhang L et al (2020) HDAC6-mediated alpha-tubulin deacetylation suppresses autophagy and enhances motility of podocytes in diabetic nephropathy. J Cell Mol Med 24(19):11558–11572
Hong Q, Zhang L, Das B, Li Z, Liu B, Cai G et al (2018) Increased podocyte Sirtuin-1 function attenuates diabetic kidney injury. Kidney Int 93(6):1330–1343
Menini S, Iacobini C, Oddi G, Ricci C, Simonelli P, Fallucca S et al (2007) Increased glomerular cell (podocyte) apoptosis in rats with streptozotocin-induced diabetes mellitus: role in the development of diabetic glomerular disease. Diabetologia 50(12):2591–2599
Kim EJ, Kho JH, Kang MR, Um SJ (2007) Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell 28(2):277–290
Wakino S, Hasegawa K, Itoh H (2015) Sirtuin and metabolic kidney disease. Kidney Int 88(4):691–698
Hasegawa K, Wakino S, Simic P, Sakamaki Y, Minakuchi H, Fujimura K et al (2013) Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nat Med 19(11):1496–1504
Liu M, Liang K, Zhen J, Zhou M, Wang X, Wang Z et al (2017) Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat Commun 8(1):413
Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS (2004) Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res 64(3):1079–1086
Van Beneden K, Geers C, Pauwels M, Mannaerts I, Verbeelen D, van Grunsven LA et al (2011) Valproic acid attenuates proteinuria and kidney injury. J Am Soc Nephrol 22(10):1863–1875
Khan S, Jena G, Tikoo K, Kumar V (2015) Valproate attenuates the proteinuria, podocyte and renal injury by facilitating autophagy and inactivation of NF-kappaB/iNOS signaling in diabetic rat. Biochimie 110:1–16
Gilbert RE, Huang Q, Thai K, Advani SL, Lee K, Yuen DA et al (2011) Histone deacetylase inhibition attenuates diabetes-associated kidney growth: potential role for epigenetic modification of the epidermal growth factor receptor. Kidney Int 79(12):1312–1321
Xue H, Li P, Luo Y, Wu C, Liu Y, Qin X et al (2019) Salidroside stimulates the Sirt1/PGC-1alpha axis and ameliorates diabetic nephropathy in mice. Phytomedicine 54:240–247
Zhang T, Chi Y, Kang Y, Lu H, Niu H, Liu W et al (2019) Resveratrol ameliorates podocyte damage in diabetic mice via SIRT1/PGC-1alpha mediated attenuation of mitochondrial oxidative stress. J Cell Physiol 234(4):5033–5043
Wang X, Gao Y, Tian N, Wang T, Shi Y, Xu J et al (2019) Astragaloside IV inhibits glucose-induced epithelial-mesenchymal transition of podocytes through autophagy enhancement via the SIRT-NF-kappaB p65 axis. Sci Rep 9(1):323
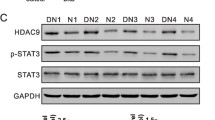
Liu F, Zong M, Wen X, Li X, Wang J, Wang Y et al (2016) Silencing of histone deacetylase 9 expression in podocytes attenuates kidney injury in diabetic nephropathy. Sci Rep 6:33676
Hassa PO, Haenni SS, Elser M, Hottiger MO (2006) Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70(3):789–829
Messner S, Hottiger MO (2011) Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol 21(9):534–542
Jason LJ, Moore SC, Lewis JD, Lindsey G, Ausio J (2002) Histone ubiquitination: a tagging tail unfolds? BioEssays 24(2):166–174
Uckelmann M, Sixma TK (2017) Histone ubiquitination in the DNA damage response. DNA Repair (Amst) 56:92–101
Goru SK, Gaikwad AB (2018) Novel reno-protective mechanism of Aspirin involves H2AK119 monoubiquitination and Set7 in preventing type 1 diabetic nephropathy. Pharmacol Rep 70(3):497–502
Khalil AM, Wahlestedt C (2008) Epigenetic mechanisms of gene regulation during mammalian spermatogenesis. Epigenetics-Us 3(1):21–28
Rossetto D, Avvakumov N, Cote J (2012) Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics-Us 7(10):1098–1108
Zhang T, Cooper S, Brockdorff N (2015) The interplay of histone modifications - writers that read. Embo Rep 16(11):1467–1481
Cruickshank MN, Besant P, Ulgiati D (2010) The impact of histone post-translational modifications on developmental gene regulation. Amino Acids 39(5):1087–1105
Hammond SL, Byrum SD, Namjoshi S, Graves HK, Dennehey BK, Tackett AJ et al (2014) Mitotic phosphorylation of histone H3 threonine 80. Cell Cycle 13(3):440–452
Park CH, Kim KT (2012) Apoptotic phosphorylation of histone H3 on Ser-10 by protein kinase Cdelta. PLoS ONE 7(9):e44307
Zhao H, Huang X, Halicka HD, Darzynkiewicz Z (2019) Detection of histone H2AX phosphorylation on Ser-139 as an indicator of DNA damage. Curr Protoc Cytom 89(1):e55
Kuo LJ, Yang LX (2008) Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo 22(3):305–309
Alghamdi TA, Batchu SN, Hadden MJ, Yerra VG, Liu Y, Bowskill BB et al (2018) Histone H3 serine 10 phosphorylation facilitates endothelial activation in diabetic kidney disease. Diabetes 67(12):2668–2681
Liao JK (2013) Linking endothelial dysfunction with endothelial cell activation. J Clin Invest 123(2):540–541
Navarro-Gonzalez JF, Mora-Fernandez C, Muros DFM, Garcia-Perez J (2011) Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat Rev Nephrol 7(6):327–340
Khan DH, Healy S, He S, Lichtensztejn D, Klewes L, Sharma KL et al (2017) Mitogen-induced distinct epialleles are phosphorylated at either H3S10 or H3S28, depending on H3K27 acetylation. Mol Biol Cell 28(6):817–824
Li K, Wang Z (2021) Histone crotonylation-centric gene regulation. Epigenetics Chromatin 14(1):10
Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E et al (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146(6):1016–1028
Martinez-Moreno JM, Fontecha-Barriuso M, Martin-Sanchez D, Sanchez-Nino MD, Ruiz-Ortega M, Sanz AB et al (2020) The contribution of histone crotonylation to tissue health and disease: focus on kidney health. Front Pharmacol 11:393
Poplawski MM, Mastaitis JW, Isoda F, Grosjean F, Zheng F, Mobbs CV (2011) Reversal of diabetic nephropathy by a ketogenic diet. PLoS ONE 6(4):e18604
Luo W, Yu Y, Wang H, Liu K, Wang Y, Huang M et al (2020) Up-regulation of MMP-2 by histone H3K9 beta-hydroxybutyrylation to antagonize glomerulosclerosis in diabetic rat. Acta Diabetol 57(12):1501–1509
Chang B, Chen Y, Zhao Y, Bruick RK (2007) JMJD6 is a histone arginine demethylase. Science 318(5849):444–447
Duan R, Ryu HY, Ahn SH (2020) Symmetric dimethylation on histone H4R3 associates with histone deacetylation to maintain properly polarized cell growth. Res Microbiol 171(2):91–98
Bouchard C, Sahu P, Meixner M, Notzold RR, Rust MB, Kremmer E et al (2018) Genomic location of PRMT6-dependent H3R2 methylation is linked to the transcriptional outcome of associated genes. Cell Rep 24(12):3339–3352
Li HT, Gong T, Zhou Z, Liu YT, Cao X, He Y et al (2015) Yeast Hmt1 catalyses asymmetric dimethylation of histone H3 arginine 2 in vitro. Biochem J 467(3):507–515
Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S et al (2011) Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc Natl Acad Sci U S A 108(51):20538–20543
Collins BE, Greer CB, Coleman BC, Sweatt JD (2019) Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics Chromatin 12(1):7
Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24(21):9630–9645
Wesche J, Kuhn S, Kessler BM, Salton M, Wolf A (2017) Protein arginine methylation: a prominent modification and its demethylation. Cell Mol Life Sci 74(18):3305–3315
Zhang J, Jing L, Li M, He L, Guo Z (2019) Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review). Mol Med Rep 19(5):3963–3971
Jiang Y, Li C, Wu Q, An P, Huang L, Wang J et al (2019) Iron-dependent histone 3 lysine 9 demethylation controls B cell proliferation and humoral immune responses. Nat Commun 10(1):2935
Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L et al (2019) H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363(6424):294–297
Wang Y, Chen W, Lian J, Zhang H, Yu B, Zhang M et al (2020) The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death Differ 27(2):695–710
Liu M, Zhang Q, Pei L, Zou Y, Chen G, Wang H (2019) Corticosterone rather than ethanol epigenetic programmed testicular dysplasia caused by prenatal ethanol exposure in male offspring rats. Epigenetics-Us 14(3):245–259
Li X, Chen X, Zhou W, Ji S, Li X, Li G et al (2017) Effect of melatonin on neuronal differentiation requires CBP/p300-mediated acetylation of histone H3 lysine 14. Neuroscience 364:45–59
Newman DM, Voss AK, Thomas T, Allan RS (2017) Essential role for the histone acetyltransferase KAT7 in T cell development, fitness, and survival. J Leukoc Biol 101(4):887–892
Maltby VE, Martin BJ, Brind’Amour J, Chruscicki AT, Mcburney KL, Schulze JM et al (2012) Histone H3K4 demethylation is negatively regulated by histone H3 acetylation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 109(45):18505–18510
Cheng SL, Ramachandran B, Behrmann A, Shao JS, Mead M, Smith C et al (2015) Vascular smooth muscle LRP6 limits arteriosclerotic calcification in diabetic LDLR-/- mice by restraining noncanonical Wnt signals. Circ Res 117(2):142–156
Henry RA, Kuo YM, Andrews AJ (2013) Differences in specificity and selectivity between CBP and p300 acetylation of histone H3 and H3/H4. Biochemistry-Us 52(34):5746–5759
Seto E, Yoshida M (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6(4):a18713
Poulard C, Corbo L, Le Romancer M (2016) Protein arginine methylation/demethylation and cancer. Oncotarget 7(41):67532–67550
Xiao J, Zhang H, Xing L, Xu S, Liu H, Chong K et al (2013) Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J Plant Physiol 170(4):444–451
Li F, Wu R, Cui X, Zha L, Yu L, Shi H et al (2016) Histone deacetylase 1 (HDAC1) negatively regulates thermogenic program in brown adipocytes via coordinated regulation of histone H3 lysine 27 (H3K27) deacetylation and methylation. J Biol Chem 291(9):4523–4536
Caslini C, Hong S, Ban YJ, Chen XS, Ince TA (2019) HDAC7 regulates histone 3 lysine 27 acetylation and transcriptional activity at super-enhancer-associated genes in breast cancer stem cells. Oncogene 38(39):6599–6614
Skucha A, Ebner J, Grebien F (2019) Roles of SETD2 in leukemia-transcription, DNA-damage, and beyond. Int J Mol Sci 20(5)
Kang JY, Kim JY, Kim KB, Park JW, Cho H, Hahm JY et al (2018) KDM2B is a histone H3K79 demethylase and induces transcriptional repression via sirtuin-1-mediated chromatin silencing. Faseb J 32(10):5737–5750
Li S, Ali S, Duan X, Liu S, Du J, Liu C et al (2018) JMJD1B Demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep 23(2):389–403
Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos AF et al (2012) Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics-Us 7(4):340–343
Ferreira RC, Popova EY, James J, Briones MR, Zhang SS, Barnstable CJ (2017) Histone deacetylase 1 is essential for rod photoreceptor differentiation by regulating acetylation at histone H3 lysine 9 and histone H4 lysine 12 in the mouse retina. J Biol Chem 292(6):2422–2440
Metzger E, Wang S, Urban S, Willmann D, Schmidt A, Offermann A et al (2019) KMT9 monomethylates histone H4 lysine 12 and controls proliferation of prostate cancer cells. Nat Struct Mol Biol 26(5):361–371
Shen H, Xu W, Lan F (2017) Histone lysine demethylases in mammalian embryonic development. Exp Mol Med 49(4):e325
Acknowledgements
The authors would like to thank the anonymous reviewers for their helpful remarks. They also thank the associate editor and the reviewers for their useful feedback that improved this paper.
Author information
Authors and Affiliations
Contributions
All authors contributed to the review conception and design. Literature reading, associated manuscript collection, and analysis were performed by Simeng Wang, Xinyu Zhang, and Qinglian Wang. The modification of this review was performed by Simeng Wang, Qinglian Wang, and Rong Wang. The first draft of the manuscript was written by Simeng Wang, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, S., Zhang, X., Wang, Q. et al. Histone modification in podocyte injury of diabetic nephropathy. J Mol Med 100, 1373–1386 (2022). https://doi.org/10.1007/s00109-022-02247-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-022-02247-7