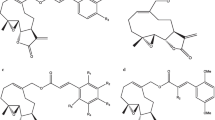
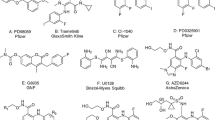
Abstract
Maternal embryonic leucine zipper kinase (MELK) is a serine-threonine kinase. Several studies have revealed its role as a regulator in the tumorigenesis of various cancers. Consequently, MELK has been considered as an attractive therapeutic target for cancer management. Herein, we report pharmacophore models extracted from crystallographic complexes of potent ligands bound to MELK protein. The resulting models were evaluated by Receiver Operating Characteristic (ROC) analysis, and the best models were employed as search queries to screen the national cancer institute for potent inhibitors. The anti-MELK bioactivities of acquired hits were evaluated in vitro. Moreover, best anti-MELK hits were further evaluated against lung and cervical cancer cells (A549 and HeLa, respectively) using cell viability MTT assay. The enzymatic assay identified six potent hits with IC50 values ranging from nanomolar to low micromolar values. The most active hit showed anti-MELK IC50 of 134.6 nM. Likewise, these hits significantly inhibited the growth of tested cancer cell lines. Interestingly, four of the identified inhibitors have chemical scaffolds that are notably different from those of reported MELK inhibitors. This study highlights the use of X-ray crystallographic structures to boost the drug discovery process.








Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Tang Q, Li W, Zheng X, Ren L, Liu J, Li S, et al. MELK is an oncogenic kinase essential for metastasis, mitotic progression, and programmed death in lung carcinoma. Signal Transduct Target Ther. 2020;5:279.
Das A, Prajapati A, Karna A, Sharma HK, Uppal S, Lather V, et al. Structure-based virtual screening of chemical libraries as potential MELK inhibitors and their therapeutic evaluation against breast cancer. Chem Biol Interact. 2023;376:110443.
Ren L, Guo J-S, Li Y-H, Dong G, Li X-Y. Structural classification of MELK inhibitors and prospects for the treatment of tumor resistance: a review. Biomed Pharmacother. 2022;156:113965.
Ganguly R, Mohyeldin A, Thiel J, Kornblum HI, Beullens M, Nakano I. MELK—a conserved kinase: functions, signaling, cancer, and controversy. Clin Transl Med. 2015;4:1–8.
Jiang P, Zhang D. Maternal embryonic leucine zipper kinase (MELK): a novel regulator in cell cycle control, embryonic development, and cancer. Int J Mol Sci. 2013;14:21551–60.
Yang H, Zhou H, Wang G, Tian L, Li H, Zhang Y, et al. MELK is a prognostic biomarker and correlated with immune infiltration in glioma. Front Neurol. 2022;13:977180–95.
Sun H, Ma H, Zhang H, Ji M. Up-regulation of MELK by E2F1 promotes the proliferation in cervical cancer cells. Int J Biol Sci. 2021;17:3875.
Li G, Yang M, Zuo L, Wang MX. MELK as a potential target to control cell proliferation in triple‑negative breast cancer MDA‑MB‑231 cells. Oncol Lett. 2018;15:9934–40.
Talib WH, Alsayed AR, Barakat M, Abu-Taha MI, Mahmod AI. Targeting drug chemo-resistance in cancer using natural products. Biomedicines 2021;9:1353.
Zhang Y, Zhou X, Li Y, Xu Y, Lu K, Li P, et al. Inhibition of maternal embryonic leucine zipper kinase with OTSSP167 displays potent anti-leukemic effects in chronic lymphocytic leukemia. Oncogene. 2018;37:5520–33.
Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig P-A, et al. The target landscape of clinical kinase drugs. Science. 2017;358:eaan4368.
Ji W, Arnst C, Tipton AR, Bekier ME, Taylor WR, Yen TJ, et al. OTSSP167 abrogates mitotic checkpoint through inhibiting multiple mitotic kinases. PLoS One. 2016;15:e0153518.
Chung S, Suzuki H, Miyamoto T, Takamatsu N, Tatsuguchi A, Ueda K, et al. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget. 2012;3:1629.
Kapale SS, Mali SN, Chaudhari HK. Molecular modelling studies for 4-oxo-1, 4-dihydroquinoline-3-carboxamide derivatives as anticancer agents. Med Drug Discov. 2019;2:100008.
Siju E, Rajalakshmi G, Paulose AP, Dhanya F, Hariraj N, Rahul K CADD: pharmacological approaches in drug design and drug discovery. World J Pharm Pharm Sci. 2017;892–908.
Gao Q, Yang L, Zhu Y. Pharmacophore-based drug design approach as a practical process in drug discovery. Curr Comput Aided Drug Des. 2010;6:37–49.
Abuhammad A, Taha M. Innovative computer-aided methods for the discovery of new kinase ligands. Future Med Chem. 2016;8:509–26.
Aparoy P, Kumar Reddy K, Reddanna P. Structure and ligand-based drug design strategies in the development of novel 5-LOX inhibitors. Curr Med Chem. 2012;19:3763–78.
Al-Sha’er MA, Mansi I, Khanfar M, Abudayyh A. Discovery of new heat shock protein 90 inhibitors using virtual co-crystallized pharmacophore generation. J Enzym Inhib Med Chem. 2016;31:64–77.
Böhm H-J, Flohr A, Stahl M. Scaffold hopping. Drug Discov Today Technol. 2004;1:217–24.
Hu Y, Stumpfe D, Bajorath J, Jr.Recent advances in scaffold hopping.J Med Chem. 2017;60:1238–46.
Yang S-Y. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today Technol. 2010;15:444–50.
Hessler G, Baringhaus K-H. The scaffold hopping potential of pharmacophores. Drug Discov Today Technol. 2010;7:e263–e269.
Zhou S, Li GB, Luo L, Zhong L, Chen K, Li H, et al. Structure-based discovery of new maternal embryonic leucine zipper kinase inhibitors. Org Biomol Chem. 2018;16:1489–95.
Carvalho AL, Trincão J, Romão MJ. X-ray crystallography in drug discovery. Methods Mol Biol. 2009;572:31–56.
Zheng H, Hou J, Zimmerman MD, Wlodawer A, Minor W. The future of crystallography in drug discovery. Expert Opin Drug Discov. 2014;9:125–37.
Triballeau N, Acher F, Brabet I, Pin J-P, Bertrand H-O. Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J Med Chem. 2005;48:2534–47.
Daoud S, Taha M. Ligand-based modeling of CXC chemokine receptor 4 and identification of inhibitors of novel chemotypes as potential leads towards new anti-COVID-19 treatments. Med Chem. 2022;18:871–83.
Irwin JJ, Shoichet BK. ZINC− a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–82.
Kirchmair J, Distinto S, Markt P, Schuster D, Spitzer GM, Liedl KR, et al. How to optimize shape-based virtual screening: choosing the right query and including chemical information. J Chem Inf Model 2009;49:678–92.
Kirchmair J, Markt P, Distinto S, Wolber G, Langer T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection—what can we learn from earlier mistakes? J Comput Aided Mol Des. 2008;22:213–28.
Al-Tawil MF, Daoud S, Ma’mon MH, Taha MO. Discovery of new Cdc2-like kinase 4 (CLK4) inhibitors via pharmacophore exploration combined with flexible docking-based ligand/receptor contact fingerprints and machine learning. RSC Adv. 2022;12:10686–700.
Shoichet BK. Interpreting steep dose-response curves in early inhibitor discovery. J Med Chem. 2006;49:7274–7.
Mousa LA, Hatmal MMM, Taha M. Exploiting activity cliffs for building pharmacophore models and comparison with other pharmacophore generation methods: sphingosine kinase 1 as case study. J Comput Aided Mol Des. 2022;36:39–62.
Sjöstedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367:eaay5947.
Kelder J, Grootenhuis PD, Bayada DM, Delbressine LP, Ploemen JP. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res. 1999;16:1514–9.
Berman H, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Mol Biol. 2003;10:980.
Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45:160–9.
Mansi IA, Al-Sha’er MA, Mhaidat NM, Taha MO, Shahin R. Investigation of binding characteristics of Phosphoinositide-dependent kinase-1 (PDK1) co-crystallized ligands through virtual pharmacophore modeling leading to novel anti-PDK1 hits. Med Chem. 2020;16:860–80.
Al-Sha’er MA, Mansi I, Almazari I, Hakooz N. Evaluation of novel Akt1 inhibitors as anticancer agents using virtual co-crystallized pharmacophore generation. J Mol Graph Model. 2015;62:213–25.
Lipinski CA. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–41.
Antony J, Tkachenko V, Lipinski C, Ekins S. Free online resources enabling crowd-sourced drug discovery. Drug Discov Today Technol. 2009;10:33.
Ma H, Deacon S, Horiuchi K. The challenge of selecting protein kinase assays for lead discovery optimization. Expert Opin Drug Discov. 2008;3:607–21.
Bardaweel S, Aljanabi R, Sabbah D, Sweidan K. Design, synthesis, and biological evaluation of novel MAO-A inhibitors targeting lung cancer. Molecules. 2022;27:2887.
Bardaweel SK, Tawaha KA, Hudaib MM. Antioxidant, antimicrobial and antiproliferative activities of Anthemis palestina essential oil. BMC Complement Altern Med. 2014;14:1–8.
Rahal BA, Bardaweel SK. Implications and efficacy of aromatase inhibitors in combination and monotherapy for the treatment of lung cancer. Anticancer Agents Med Chem. 2022;22:3114–24.
Acknowledgements
The authors thank the Deanships of Scientific Research at the Applied Sciences Private University (Amman) and University of Jordan for sponsoring this project.
Funding
This work was supported by the Deanships of Scientific Research at the Applied Sciences Private University (Amman) and University of Jordan.
Author information
Authors and Affiliations
Contributions
SD: Investigation, Formal analysis, Writing, Review & Editing. SJA: Formal analysis, Review & Editing. SB: Formal analysis, Review & Editing. MOT Conceptualization, Methodology, Supervision, Investigation, Resources, Writing, Review & Editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Daoud, S., Alabed, S.J., Bardaweel, S.K. et al. Discovery of potent maternal embryonic leucine zipper kinase (MELK) inhibitors of novel chemotypes using structure-based pharmacophores. Med Chem Res 32, 2574–2586 (2023). https://doi.org/10.1007/s00044-023-03160-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-023-03160-5