Abstract
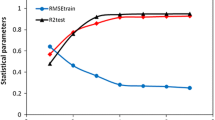
A quantitative structure–activity relationship (QSAR) model has been carried out for a series of 3-acylamino-2-aminopropionic acid derivatives with high affinities in a binding assay for the glycine site. The replacement method (RM) and stepwise-multiple linear regression (Stepwise-MLR) strategies are used as feature selection (descriptor selection). The model of the relationship between selected molecular descriptors and pK i data has been achieved by linear (multiple linear regression, MLR) and nonlinear (support vector machine, SVM) methods. Leave-one-out cross-validation and external-validation were carried out with the aim for evaluating the predictive capability of the models. The squared correlation coefficients of experimental versus predicted activities for the test set obtained by MLR and SVM models using RM feature selection are 0.463 and 0.655, respectively. Our best QSAR model illustrates the importance of an adequate distribution of atomic properties represented in 3D frames and reveals the mass and van der Waals volumes as the most influencing atomic properties in the structures of the 3-acylamino-2-aminopropionic acid derivatives.



Similar content being viewed by others
References
Abreu RMV, Ferreira ICFR, Queiroz MJRP (2009) QSAR model for predicting radical scavenging activity of di(hetero)arylamines derivatives of benzo[b]thiophenes. Eur J Med Chem 44:1952–1958
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Control 19:716–723
Balsamini C, Bedini A, Diamantini G, Spadoni G, Tontini A, Tarzia G (1998) (E)-3-(2-(N-phenylcarbamoyl)vinyl)pyrrole-2-carboxylic acid derivatives. A novel class of glycine site antagonists. J Med Chem 41:808–820
Bare TM (1998) Pyridazino[4,5-b]quinolinediones: novel glycine/N-methyl-d-aspartate antagonists for the treatment of stroke. J Heterocycl Chem 35:1171–1186
Brereton RG (2007) Applied chemometrics for scientists. Wiley, New York
Buchstaller HP, Siebert CD, Steinmetz R, Frank I, Berger ML, Gottschlich R, Leibrock J, Krug M, Steinhilber D, Noe CR (2006) Synthesis of thieno[2,3-b]pyridinones acting as cytoprotectants and as inhibitors of [3H]glycine binding to the N-methyl-d-aspartate (NMDA) receptor. J Med Chem 49:864–871
Buyukbingol E, Sisman A, Akyildiz M, Alparslan FN, Adejare A (2007) Adaptive neuro-fuzzy inference system (ANFIS): a new approach to predictive modeling in QSAR applications: a study of neuro-fuzzy modeling of PCP-based NMDA receptor antagonists. Bioorg Med Chem 15:4265–4282
Carling RW, Leeson PD, Moore KW, Moyes CR, Duncton M, Hudson ML, Baker R, Foster AC, Grimwood S, Kemp JA, Marshall GR, Tricklebank MD, Saywell KL (1997) 4-Substituted-3-phenylquinolin-2(1H)-ones: acidic and nonacidic Glycine site N-Methyl-d-aspartate antagonists with in vivo activity. J Med Chem 40:754–765
Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, Tong G, Lipton SA, Zhang D (2002) Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature 415:793–798
Cheng ZJ, Zhang YT, Zhang WJ (2009a) QSAR studies of imidazopyridine derivatives as Et-PKG inhibitors by using PSO-SVM approach. Med Chem Res. doi:10.1007/s00044-009-9272-y
Cheng ZJ, Zhang YT, Zhou CH, Zhang WJ, Gao SB (2009b) Classification of 5-HT1A receptor ligands on the basis of their binding affinities by using PSO-Adaboost-SVM. Int J Mol Sci 10:3316–3337
Cherqaoui D, Esseffar M, Villemin D, Cense JM, Chastrette M, Zakarya D (1998) Structure-musk odour relationship studies of tetralin and indan compounds using neural networks. New J Chem 22:839–843
Chihchung C, Chihjen L (2009) LIBSVM-a library for support vector machines. http://www.csie.ntu.edu.tw/~cjlin/libsvm
Clementi S, Wold S (1995) How to choose the proper statistical method. In: van de Waterbeemd H (ed) Chemometrics methods in molecular design. VCH, Weinheim, pp 319–338
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Dougherty ER, Barrera J, Brun M, Kim S, Cesar RM, Chen Y, Bittner M, Trent JM (2002) Inference from clustering with application to gene-expression microarrays. J Comput Biol 9:105–126
Doytchinova IA, Walshe V, Borrow P, Flower DR (2005) Towards the chemometric dissection of peptide—HLA-A*0201 binding affinity: comparison of local and global QSAR models. J Comput Aid Mol Des 15:203–212
Duchowicz PR, Fernández FM, Castro EA (2006) Alternative algorithm for the search of an optimal set of descriptors in QSAR-QSPR studies. MATCH Commun Math Comput Chem 55:179–192
Duchowicz PR, González MP, Helguera AM, Cordeiro MDNS, Castro EA (2007) Application of the replacement method as novel variable selection in QSPR. 2. Soil sorption coefficients. Chemom Intell Lab Syst 88:197–203
González MP, Caballero J, Camba AT, Helguerab AM, Fernándezc M (2006a) Modeling of farnesyltransferase inhibition by some thiol and non-thiol peptidomimetic inhibitors using genetic neural networks and RDF approaches. Bioorgan Med Chem 14:200–213
González MP, Puente M, Fall Y, Gómez G (2006b) In silico studies using radial distribution function approach for predicting affinity of 1α,25-dihydroxyvitamin D3 analogues for Vitamin D receptor. Steroids 71:510–527
Goodarzi M, Freitas MP (2008) Augmented three-mode MIA-QSAR modeling for a series of anti-HIV-1 compounds. QSAR Comb Sci 27:1092–1098
Huettner JE (1991) Competitive antagonism of glycine at the N-methyl-d-aspartate (NMDA) receptor. Biochem Pharmacol 41:9–16
Jansen M, Dannhardt G (2003) Antagonists and agonists at the glycine site of the NMDA receptor for therapeutic interventions. Eur J Med Chem 38:661–670
Johnson JW, Ascher P (1987) Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature 325:529–531
Kemp JA, McKernan RM (2002) NMDA receptor pathways as drug targets. Nat Neurosci 5:1039–1042
Kleckner NW, Dingledine R (1988) Requirements for glycine in activation of NMDA receptors expressed in Xenopus oocytes. Science 241:835–837
Kubinyi H (1994a) Variable selection in QSAR studies. I. An evolutionary algorithm. Quant Struct Act Relatsh 13:285–294
Kubinyi H (1994b) Variable selection in QSAR studies. II. A highly efficient combination of systematic search and evolution. Quant Struct Act Relatsh 13:393–401
Kulagowski JJ (1996) Glycine-site NMDA receptor antagonists: an update. Exp Opin Ther Pat 6:1069–1079
Kulagowski JJ, Leeson PD (1995) Glycine-site NMDA receptor antagonists. Exp Opin Ther Pat 5:1061–1075
Labrie V, Roder JC (2010) The involvement of the NMDA receptor d-serine/glycine site in the pathophysiology and treatment of schizophrenia. Neurosci Biobehav Rev 34:351–372
Leeson PD, Iversen LL (1994) The glycine site on the NMDA receptor: structure–activity relationship and therapeutic potential. J Med Chem 37:4053–4067
Li XM, Zhao M, Tang YR, Wang C, Zhang ZD, Peng SQ (2008) N-[2-(5,5-Dimethyl-1,3-dioxane-2-yl)ethyl]amino acids: their synthesis, anti-inflammatory evaluation and QSAR analysis. Eur J Med Chem 43:8–18
Liu HX, Zhang RS, Yao XJ, Liu MC, Hul ZD, Fan BT (2004) QSAR and classification models of a novel series of COX-2 selective inhibitors: 1,5-diarylimidazoles based on support vector machines. J Comput Aid Mol Des 18:389–399
Mercader AG, Duchowicz PR, Fernández FM, Castro EA (2008) Modified and enhanced replacement method for the selection of molecular descriptors in QSAR and QSPR theories. Chemom Intell Lab Syst 92:138–144
Newcomer JW, Krystal JH (2001) NMDA receptor regulation of memory and behavior in humans. Hippocampus 11:529–542
Panek JJ, Jezierska A, Vračko M (2005) Kohonen network study of aromatic compounds based on electronic and nonelectronic structure descriptors. J Chem Inf Model 45:264–272
Schneider CI, Urwyler S (1992) Biochemical and thermodynamic aspects of the binding of [3H]glycine to its strychnine-insensitive recognition site associated with the N-methyl-d-aspartate receptor complex. Biochem Pharmacol 43:1693–1699
Sia HZ, Zhang KJ, Hua ZD, Fand BT (2007) QSAR model for prediction capacity factor of molecular imprinting polymer based on gene expression programming. QSAR Comb Sci 26:41–50
Sills MA, Fagg G, Pozza M, Angst C, Brundish DE, Hurt SD, Wilusz EJ, Williams M (1991) [3H]CGP39653: a new N-methyl-d-aspartate antagonist radioligand with low nanomolar affinity in rat brain. Eur J Pharmacol 192:19–24
Tedesco G, Feriani A, Mor M (2000) Statistical analysis on a series of glycine antagonists. II Farmaco 55:194–196
Thomson AM (1990) Glycine is a coagonist at the NMDA receptor/ion channel complex. Progr Neurobiol 35:53–74
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley-VCH, Weinheim (Germany)
Urwyler S, Laurie D, Lowe DA, Meier CL, Muller W (1996) Biphenyl-derivatives of 2-amino-7-phosphonoheptanoic acid, a novel class of potent competitive N-methyl-d-aspartate receptor antagonists. I. Pharmacological characterization in vitro. Neuropharmacology 35:643–654
Urwyler S, Floersheim P, Roy BL, Koller M (2009) Drug design, in vitro pharmacology, and structure–activity relationships of 3-acylamino-2-aminopropionic acid derivatives, a novel class of partial Agonists at the glycine site on the N-Methyl-d-aspartate (NMDA) receptor complex. J Med Chem 52:5093–5107
Yu YJ, Su RX, Wang LB, Qi W, He ZM (2009) Comparative QSAR modeling of antitumor activity of ARC-111 analogues using stepwise MLR, PLS, and ANN techniques. Med Chem Res. doi:10.1007/s00044-009-9266-9
Yuan YN, Zhang RS, Hu RJ, Ruan XF (2009) Prediction of CCR5 receptor binding affinity of substituted 1-(3,3-diphenylpropyl)-piperidinyl amides and ureas based on the heuristic method, support vector machine and projection pursuit regression. Eur J Med Chem 44:25–34
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, Z., Zhang, Y. & Fu, W. Predictive QSAR models of 3-acylamino-2-aminopropionic acid derivatives as partial agonists of the glycine site on the NMDA receptor. Med Chem Res 20, 1235–1246 (2011). https://doi.org/10.1007/s00044-010-9464-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-010-9464-5