Abstract
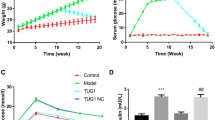
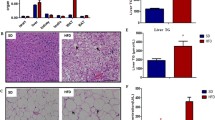
White adipose tissue (WAT) is important for regulating the whole systemic energy homeostasis. Excessive WAT accumulation further contributes to the development of obesity and obesity-related illnesses. More detailed mechanisms for WAT lipid metabolism reprogramming, however, are still elusive. Here, we report the abnormally high expression of a circular RNA (circRNA) mmu_circ_0001874 in the WAT and liver of mice with obesity. mmu_circ_0001874 interference achieved using a specific adeno-associated virus infects target tissues, down-regulating lipid accumulation in the obesity mice WAT, and liver tissues. Mechanistically, miR-24-3p directly interacts with the lipid metabolism effect of mmu_circ_0001874 and participates in adipogenesis and lipid accumulation by targeting Igf2/PI3K-AKT-mTOR axis. Moreover, mmu_circ_0001874 binds to Igf2bp2 to interact with Ucp1, up-regulating Ucp1 translation and increasing thermogenesis to decrease lipid accumulation. In conclusion, our data highlight a physiological role for circRNA in lipid metabolism reprogramming and suggest mmu_circ_0001874/miR-24-3p/Igf2/PI3K-AKT-mTOR and mmu_circ_0001874/Igf2bp2/Ucp1 axis may represent a potential mechanism for controlling lipid accumulation in obesity.









Similar content being viewed by others
Availability of data and materials
The sequencing data are available on NCBI database at Sequence Read Archive (SRA). The transcriptome sequencing data: SUB12222924.
References
Ma Y, Li J, Ju Z, Huang W, Wang Z, Yang L, Ding L (2021) Danning tablets alleviate high fat diet-induced obesity and fatty liver in mice via modulating SREBP pathway. J Ethnopharmacol 279:114320
Ramos-Arellano L, Matia-Garcia I, Marino-Ortega L, Castro-Alarcón N, Muñoz-Valle J, Salgado-Goytia L, Salgado-Bernabé A, Parra-Rojas I (2020) Obesity, dyslipidemia, and high blood pressure are associated with cardiovascular risk, determined using high-sensitivity C-reactive protein concentration, in young adults. J Int Med Res 48:300060520980596
Brown J, Yang S, Mire E, Wu X, Miele L, Ochoa A, Zabaleta J, Katzmarzyk P (2021) Obesity and cancer risk in white and black adults: a prospective cohort study. Obesity 29:960–965
Cooper C, Mandel E (2020) Recognizing and treating child overweight and obesity. JAAPA 33:47–50
Llewellyn C, Wardle J (2015) Behavioral susceptibility to obesity: Gene–environment interplay in the development of weight. Physiol Behav 152:494–501
Swinburn B, Sacks G, Hall K, McPherson K, Finegood D, Moodie M, Gortmaker S (2011) The global obesity pandemic: shaped by global drivers and local environments. Lancet 378:804–818
Yu G, Yang Z, Peng T, Lv Y (2021) Circular RNAs: Rising stars in lipid metabolism and lipid disorders. J Cell Physiol 236:4797–4806
Jeyakumar S, Yasmeen R, Reichert B, Ziouzenkova O (2013) Metabolism of vitamin A in white adipose tissue and obesity. Carotenoids and vitamin A in translational medicine. CRC Press, Boca Raton, pp 23–54
Zhu Q, Scherer P (2017) Immunologic and endocrine functions of adipose tissue: implications for kidney disease. Nat Rev Nephrol 14:105–120
Maury E, Brichard S (2010) Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol Cell Endocrinol 314:1–16
Buechler C, Wanninger J, Neumeier M (2011) Adiponectin, a key adipokine in obesity related liver diseases. World J Gastroenterol 17:2801–2811
Chen X, Li HD, Bu FT, Li XF, Li J (2020) Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 10:4851–4870
Arcinas C, Tan W, Fang W, Desai T, Teh D, Degirmenci U, Xu D, Foo R, Sun L (2019) Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat Metab 1:688–703
Shi C, Huang F, Gu X, Zhang M, Wen J, Wang X, You L, Cui X, Ji C, Guo X (2016) Adipogenic miRNA and Meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget 7:40830–40845
Zhao E, Keller M, Rabaglia M, Oler A, Stapleton D, Schueler K, Neto E, Moon J, Wang P, Wang I et al (2009) Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome 20:476–485
Jiang R, Li H, Yang J, Shen X, Song C, Yang Z, Wang X, Huang Y, Lan X, Lei C (2020) circRNA profiling reveals an abundant circFUT10 that promotes adipocyte proliferation and inhibits adipocyte differentiation via sponging let-7. Mol Ther Nucl Acids 20:491–501
Ding Z, Sun D, Han J, Shen L, Yang F, Sah S, Sui X, Wu G (2021) Novel noncoding RNA CircPTK2 regulates lipolysis and adipogenesis in cachexia. Mol Metab 53:101310
Liu Y, Liu H, Li Y, Mao R, Zhang T (2020) Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 10:4705–4719
Arcinas C, Tan W, Fang W, Desai TP, Sun L (2019) Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat Metab 1:688–703
Chen Q, Liu M, Luo Y, Yu H, He Q (2020) Maternal obesity alters circRNA expression and the potential role of mmu_circRNA_0000660 via sponging miR_693 in offspring liver at weaning age. Gene 731:144354
Li P, Shan K, Liu Y, Zhang Y, Xu L, Xu L (2018) CircScd1 promotes fatty liver disease via the janus kinase 2/signal transducer and activator of transcription 5 pathway. Dig Dis Sci 64:113–122
Li J, Qi J, Tang Y, Liu H, Zhou K, Dai Z, Yuan L, Sun C (2021) A nanodrug system overexpressed circRNA_0001805 alleviates nonalcoholic fatty liver disease via miR-106a-5p/miR-320a and ABCA1/CPT1 axis. J Nanobiotechnol 19:363
Yang G, Wu M, Liu X, Wang F, Li M, An X, Bai F, Lei C, Dang R (2022) MiR-24-3p conservatively regulates muscle cell proliferation and apoptosis by targeting common gene CAMK2B in rat and cattle. Animals (Basel) 12:505
Gao Z, Zhou L, Hua S, Wu H, Luo L, Li L, Wang S, Liu Y, Zhou Z, Chen X (2020) miR-24-3p promotes colon cancer progression by targeting ING1. Signal Transduct Targ Ther 5:171
Liu L, Liu L, Lu Y, Zhang T, Zhao W (2021) Serum aberrant expression of miR-24-3p and its diagnostic value in Alzheimer’s disease. Biomark Med 15:1499–1507
Guo L, Chao X, Huang W, Li Z, Luan K, Ye M, Zhang S, Liu M, Li H, Luo W et al (2021) Whole transcriptome analysis reveals a potential regulatory mechanism of LncRNA-FNIP2/miR-24-3p/FNIP2 axis in chicken adipogenesis. Front Cell Dev Biol 9:653798
Liu H, Wang X, Wang Z, Li L (2020) Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. J Cell Physiol 235:4520–4529
Zhang J, Liu L, Xue Y, Ma Y, Liu X, Li Z, Li Z, Liu Y (2018) Endothelial monocyte-activating polypeptide-II induces BNIP3-mediated mitophagy to enhance temozolomide cytotoxicity of glioma stem cells via down-regulating MiR-24-3p. Front Mol Neurosci 11:92
Yerlikaya FH (2019) Aberrant expression of miRNA profiles in high-fat and high-sucrose fed rats. Clin Nutr Exp 27:1–8
Zhang D, Lu K, Dong Z, Jiang G, Xu W, Liu W (2014) The effect of exposure to a high-fat diet on MicroRNA expression in the liver of blunt snout bream (Megalobrama amblycephala). PLoS ONE 9:e96132
Mentzel C, Anthon C, Jacobsen M, Karlskov-Mortensen P, Bruun C, Jørgensen C, Gorodkin J, Cirera S, Fredholm M (2015) Gender and obesity specific MicroRNA expression in adipose tissue from lean and obese pigs. PLoS ONE 10:e0131650
Luo G, Hu S, Lai T, Wang J, Wang L, Lai S (2020) MiR-9-5p promotes rabbit preadipocyte differentiation by suppressing leptin gene expression. Lipids Health Dis 19:126
Takahashi K, Yamaguchi S, Shimoyama T, Seki H, Miyokawa K, Katsuta H, Tanaka T, Yoshimoto K, Ohno H, Nagamatsu S et al (2008) JNK- and IkappaB-dependent pathways regulate MCP-1 but not adiponectin release from artificially hypertrophied 3T3-L1 adipocytes preloaded with palmitate in vitro. Am J Physiol Endocrinol Metab 294:E898-909
Fang W, Jia Y, Wang P, Yang Q, Chang ZJ (2017) Identification and profiling of Cyprinus carpio microRNAs during ovary differentiation by deep sequencing. BMC Genom 18:333
Pitto L, Ripoli A, Cremisi F, Rainaldi G (2008) microRNA (interference) networks are embedded in the gene regulatory networks. Cell Cycle 7:2458–2461
Shao J, Wang J, Li Y, Elzo M, Tang T, Lai T, Ma Y, Gan M, Wang L, Jia X et al (2020) Growth, behavioural, serum biochemical and morphological changes in female rabbits fed high-fat diet. J Anim Physiol Anim Nutr (Berl) 105:345–353
Li J, Liu S, Zhou H, Qu L, Yang J (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucl Acids Res 42:92–97
Kim YJ, Si YC, Yun CH, Lee TR, Sang HK (2020) Transcriptional activation of Cidec by PPARγ2 in adipocyte. Biochem Biophys Res Commun 377:297–302
Wen J, Friedman J (2012) miR-122 regulates hepatic lipid metabolism and tumor suppression. J Clin Invest 122:2773–2776
Yu G, Yang Z, Peng T, Lv Y (2020) Circular RNAs: rising stars in lipid metabolism and lipid disorders. J Cell Physiol 236:4797–4806
Pan X, Fang Y, Li X, Yang Y, Shen H (2020) RBPsuite: RNA-protein binding sites prediction suite based on deep learning. BMC Genom 21:884
Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia G (2013) catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics 29:2928–2930
Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano MJ, Jungkamp A, Munschauer M et al (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141
Ning D, Zhao L, Wrighting D, Krmer D, Majithia A, Wang Y, Cracan V, Borges-Rivera D, Mootha V, Nahrendorf M (2015) IGF2BP2/IMP2-deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins. Cell Metab 21:609–621
Farmer S (2006) Transcriptional control of adipocyte formation. Cell Metab 4:263–273
Kirkland J, Hollenberg C, Kindler S, Gillon W (1994) Effects of age and anatomic site on preadipocyte number in rat fat depots. J Gerontol 49:31–35
Xi F, Wei C, Xu Y, Ma L, He Y, Shi X, Yang G, Yu T (1816) MicroRNA-214-3p targeting Ctnnb1 promotes 3T3-L1 preadipocyte differentiation by interfering with the Wnt/β-catenin signaling pathway. Int J Mol Sci 2019:20
Li Q, Wang N, Wei H, Li C, Wu J, Yang G (2016) miR-24-3p regulates progression of gastric mucosal lesions and suppresses proliferation and invasiveness of N87 via peroxiredoxin 6. Dig Dis Sci 61:3486–3497
Liu H, Wang X, Wang Z, Li L (2019) Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. J Cell Physiol 235:4520–4529
Cui X, Huang X, Huang M, Zhou S, Le G, Yu W, Duan M, Jiang B, Zeng J, Zhou J et al (2021) miR-24-3p obstructs the proliferation and migration of HSFs after thermal injury by targeting PPAR-β and positively regulated by NF-κB. Exp Dermatol 31:841–853
Li Z, Sun Y, Cao S, Zhang J, Wei J (2019) Downregulation of miR-24-3p promotes osteogenic differentiation of human periodontal ligament stem cells by targeting SMAD family member 5. J Cell Physiol 234:7411–7419
Bai X, Zhang P, Liu Y, Liu H, Lv L, Zhou Y (2021) TRIB3 promotes osteogenic differentiation of human adipose-derived mesenchymal stem cells levelled by post-transcriptional regulation of miR-24-3p. Chin J Dent Res 24:235–249
Lin Z, Tang Y, Li Z, Li J, Yu C, Yang C, Liu L, Wang Y, Liu Y (2022) miR-24-3p Dominates the proliferation and differentiation of chicken intramuscular preadipocytes by blocking ANXA6 expression. Genes (Basel) 13:635
Yu Y, Yang T, Ding Z, Cao Y (2022) Circ_0026579 alleviates LPS-induced WI-38 cells inflammation injury in infantile pneumonia. Innate Immun 28:37–48
Sélénou C, Brioude F, Giabicani E, Sobrier M, Netchine I (1886) IGF2: development, genetic and epigenetic abnormalities. Cells 2022:11
Halmos T, Suba I (2019) The physiological role of growth hormone and insulin-like growth factors. Orv Hetil 160:1774–1783
Qiao X, Wang L, Liu M, Tian Y, Chen T (2019) MiR-210-3p attenuates lipid accumulation and inflammation in atherosclerosis by repressing IGF2. Biosci Biotechnol Biochem 84:321–329
Kessler S, Laggai S, Van Wonterg E, Gemperlein K, Müller R, Haybaeck J, Vandenbroucke R, Ogris M, Libert C, Kiemer A (2016) Transient hepatic overexpression of insulin-like growth factor 2 induces free cholesterol and lipid droplet formation. Front Physiol 7:147
Chao W, D’Amore P (2008) IGF2: Epigenetic regulation and role in development and disease. Cytokine Growth Fact Rev 19:111–120
Bian Q, Chen B, Weng B, Chu D, Tang X, Yan S, Yin Y, Ran M (2021) circBTBD7 promotes immature porcine sertoli cell growth through modulating miR-24-3p/MAPK7 axis to inactivate p38 MAPK signaling pathway. Int J Mol Sci 22:9385
Zhang L, Wang Z, Lan D, Zhao J, Wang L, Shao X, Wang D, Wu K, Sun M, Huang X et al (2022) MicroRNA-24-3p alleviates cardiac fibrosis by suppressing cardiac fibroblasts mitophagy via downregulating PHB2. Pharmacol Res 177:106124
Zhang H, Xue S, Feng Y, Shen J, Zhao J (2020) MicroRNA-24-3p inhibition prevents cell growth of vascular smooth muscle cells by targeting Bcl-2-like protein 11. Exp Ther Med 19:2467–2474
Chen Z, Liu M, Zhang W, Deng M, Zhou Y, Li Y (2020) miR-24-3p induces human intervertebral disc degeneration by targeting insulin-like growth factor binding protein 5 and the ERK signaling pathway. Life Sci 243:117288
Liu M, Xie S, Liu W, Li J, Zhang H (2019) Mechanism of SEMA3G knockdown-mediated attenuation of high-fat diet-induced obesity. J Endocrinol 244:223–236
Lagathu C, Christodoulides C, Virtue S, Cawthorn W, Franzin C, Kimber W, Nora E, Campbell M, Medina-Gomez G, Cheyette B et al (2009) Dact1, a nutritionally regulated preadipocyte gene, controls adipogenesis by coordinating the Wnt/beta-catenin signaling network. Diabetes 58:609–619
Zhou L, Xu L, Ye J, Li D, Wang W, Li X, Wu L, Wang H, Guan F, Li P (2012) Cidea promotes hepatic steatosis by sensing dietary fatty acids. Hepatology 56:95–107
Tybl E, Shi F, Kessler S, Tierling S, Walter J, Bohle R, Wieland S, Zhang J, Tan E, Kiemer A (2011) Overexpression of the IGF2-mRNA binding protein p62 in transgenic mice induces a steatotic phenotype. J Hepatol 54:994–1001
Regué L, Minichiello L, Avruch J, Dai N (2019) Liver-specific deletion of IGF2 mRNA binding protein-2/IMP2 reduces hepatic fatty acid oxidation and increases hepatic triglyceride accumulation. J Biol Chem 294:11944–11951
Enerbäck S, Jacobsson A, Simpson E, Guerra C, Yamashita H, Harper M, Kozak L (1997) Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387:90–94
Acknowledgements
The authors are grateful to Ms. Ting Pan (WestChina-Frontier PharmaTech Co., Ltd, Chengdu, China) for the provision of hepatocytes.
Funding
The National Modern Agricultural Industrial Technology System (CARS-43-A-2) and the Key Research and Development Program of Sichuan Province (2021YFYZ0033) provided funding for our research.
Author information
Authors and Affiliations
Contributions
The project was planned and designed by JS, JW, XJ, and SL. Data were gathered and research was carried out by JS, MW, ZL, GJ, and TT. JS and AZ wrote this paper.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest to declare.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The experimental procedures were approved by the Animal Care and Use Committee from the College of Animal Science and Technology, Sichuan Agricultural University, China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shao, J., Wang, M., Zhang, A. et al. Interference of a mammalian circRNA regulates lipid metabolism reprogramming by targeting miR-24-3p/Igf2/PI3K-AKT-mTOR and Igf2bp2/Ucp1 axis. Cell. Mol. Life Sci. 80, 252 (2023). https://doi.org/10.1007/s00018-023-04899-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04899-1