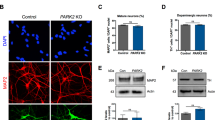
Abstract
Chediak–Higashi syndrome (CHS) is a rare, autosomal recessive disorder caused by biallelic mutations in the lysosomal trafficking regulator (LYST) gene. Even though enlarged lysosomes and/or lysosome-related organelles (LROs) are the typical cellular hallmarks of CHS, they have not been investigated in human neuronal models. Moreover, how and why the loss of LYST function causes a lysosome phenotype in cells has not been elucidated. We report that the LYST-deficient human neuronal model exhibits lysosome depletion accompanied by hyperelongated tubules extruding from enlarged autolysosomes. These results have also been recapitulated in neurons differentiated from CHS patients’ induced pluripotent stem cells (iPSCs), validating our model system. We propose that LYST ensures the correct fission/scission of the autolysosome tubules during autophagic lysosome reformation (ALR), a crucial process to restore the number of free lysosomes after autophagy. We further demonstrate that LYST is recruited to the lysosome membrane, likely to facilitate the fission of autolysosome tubules. Together, our results highlight the key role of LYST in maintaining lysosomal homeostasis following autophagy and suggest that ALR dysregulation is likely associated with the neurodegenerative CHS phenotype.






Similar content being viewed by others
Data availability
All the relevant data are within the paper and its supporting Information. Numerical data are provided in the supplementary Excel file.
Change history
02 March 2023
Typo in the author name May Christine V. Malicdan
04 March 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00018-023-04724-9
References
Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW et al (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465:942–946. https://doi.org/10.1038/nature09076
Ferguson SM (2018) Axonal transport and maturation of lysosomes. Curr Opin Neurobiol 51:45–51. https://doi.org/10.1016/j.conb.2018.02.020
Lie PPY, Nixon RA (2019) Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol Dis 122:94–105. https://doi.org/10.1016/j.nbd.2018.05.015
Winckler B, Faundez V, Maday S, Cai Q, Guimas Almeida C, Zhang H (2018) The endolysosomal system and proteostasis: from development to degeneration. J Neurosci 38:9364–9374. https://doi.org/10.1523/JNEUROSCI.1665-18.2018
Sharma P, Nicoli E-R, Serra-Vinardell J, Morimoto M, Toro C, Malicdan MCV, Introne WJ (2019) Chediak-Higashi syndrome: a review of the past, present, and future. Drug Discov Today Dis Models 31:31–36
Kaplan J, De Domenico I, Ward DM (2008) Chediak-Higashi syndrome. Curr Opin Hematol 15:22–29. https://doi.org/10.1097/MOH.0b013e3282f2bcce
Introne W, Boissy RE, Gahl WA (1999) Clinical, molecular, and cell biological aspects of Chediak-Higashi syndrome. Mol Genet Metab 68:283–303. https://doi.org/10.1006/mgme.1999.2927
Barbosa MD, Nguyen QA, Tchernev VT, Ashley JA, Detter JC, Blaydes SM, Brandt SJ, Chotai D, Hodgman C, Solari RC, Lovett M et al (1996) Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 382:262–265. https://doi.org/10.1038/382262a0
Nagle DL, Karim MA, Woolf EA, Holmgren L, Bork P, Misumi DJ, McGrail SH, Dussault BJ Jr, Perou CM, Boissy RE, Duyk GM et al (1996) Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat Genet 14:307–311. https://doi.org/10.1038/ng1196-307
Kypri E, Schmauch C, Maniak M, De Lozanne A (2007) The BEACH protein LvsB is localized on lysosomes and postlysosomes and limits their fusion with early endosomes. Traffic (Copenhagen, Denmark) 8:774–783. https://doi.org/10.1111/j.1600-0854.2007.00567.x
Perou CM, Leslie JD, Green W, Li L, Ward DM, Kaplan J (1997) The Beige/Chediak-Higashi syndrome gene encodes a widely expressed cytosolic protein. J Biol Chem 272:29790–29794. https://doi.org/10.1074/jbc.272.47.29790
Durchfort N, Verhoef S, Vaughn MB, Shrestha R, Adam D, Kaplan J, Ward DM (2012) The enlarged lysosomes in beige j cells result from decreased lysosome fission and not increased lysosome fusion. Traffic 13:108–119. https://doi.org/10.1111/j.1600-0854.2011.01300.x
Sepulveda FE, Burgess A, Heiligenstein X, Goudin N, Menager MM, Romao M, Cote M, Mahlaoui N, Fischer A, Raposo G, Menasche G et al (2015) LYST controls the biogenesis of the endosomal compartment required for secretory lysosome function. Traffic 16:191–203. https://doi.org/10.1111/tra.12244
Rudelius M, Osanger A, Kohlmann S, Augustin M, Piontek G, Heinzmann U, Jennen G, Russ A, Matiasek K, Stumm G, Schlegel J (2006) A missense mutation in the WD40 domain of murine Lyst is linked to severe progressive Purkinje cell degeneration. Acta Neuropathol 112:267–276. https://doi.org/10.1007/s00401-006-0092-6
Hedberg-Buenz A, Dutca LM, Larson DR, Meyer KJ, Soukup DA, van der Heide CJ, Mercer HE, Wang K, Anderson MG (2019) Mouse models and strain-dependency of Chediak-Higashi syndrome-associated neurologic dysfunction. Sci Rep 9:6752. https://doi.org/10.1038/s41598-019-42159-0
Trantow CM, Hedberg-Buenz A, Iwashita S, Moore SA, Anderson MG (2010) Elevated oxidative membrane damage associated with genetic modifiers of Lyst-mutant phenotypes. PLoS Genet. 6:e1001008. https://doi.org/10.1371/journal.pgen.1001008
Wang C, Ward ME, Chen R, Liu K, Tracy TE, Chen X, Xie M, Sohn PD, Ludwig C, Meyer-Franke A, Karch CM et al (2017) Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep 9:1221–1233. https://doi.org/10.1016/j.stemcr.2017.08.019
Fernandopulle MS, Prestil R, Grunseich C, Wang C, Gan L, Ward ME (2018) Transcription factor-mediated differentiation of human iPSCs into neurons. Curr Protoc Cell Biol. 79:e51. https://doi.org/10.1002/cpcb.51
Perou CM, Kaplan J (1993) Chediak-Higashi syndrome is not due to a defect in microtubule-based lysosomal mobility. J Cell Sci 106(Pt 1):99–107. https://doi.org/10.1242/jcs.106.1.99
Wang C, Telpoukhovskaia MA, Bahr BA, Chen X, Gan L (2018) Endo-lysosomal dysfunction: a converging mechanism in neurodegenerative diseases. Curr Opin Neurobiol 48:52–58. https://doi.org/10.1016/j.conb.2017.09.005
Son JH, Shim JH, Kim KH, Ha JY, Han JY (2012) Neuronal autophagy and neurodegenerative diseases. Exp Mol Med 44:89–98. https://doi.org/10.3858/emm.2012.44.2.031
Yap CC, Digilio L, McMahon LP, Garcia ADR, Winckler B (2018) Degradation of dendritic cargos requires Rab7-dependent transport to somatic lysosomes. J Cell Biol 217:3141–159. https://doi.org/10.1083/jcb.201711039
Cheng XT, Xie YX, Zhou B, Huang N, Farfel-Becker T, Sheng ZH (2018) Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J Cell Biol 217:3127–3139. https://doi.org/10.1083/jcb.201711083
Gil-Krzewska A, Saeed MB, Oszmiana A, Fischer ER, Lagrue K, Gahl WA, Introne WJ, Coligan JE, Davis DM, Krzewski K (2017) An actin cytoskeletal barrier inhibits lytic granule release from natural killer cells in patients with Chediak-Higashi syndrome. J Allergy Clin Immunol 142:914–27.e6. https://doi.org/10.1016/j.jaci.2017.10.040
Holland P, Torgersen ML, Sandvig K, Simonsen A (2014) LYST affects lysosome size and quantity, but not trafficking or degradation through autophagy or endocytosis. Traffic 15:1390–1405. https://doi.org/10.1111/tra.12227
Burkhardt JK, Wiebel FA, Hester S, Argon Y (1993) The giant organelles in beige and Chediak-Higashi fibroblasts are derived from late endosomes and mature lysosomes. J Exp Med 178:1845–1856. https://doi.org/10.1084/jem.178.6.1845
Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y et al (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141:1146–1158. https://doi.org/10.1016/j.cell.2010.05.008
Chen CS, Chen WN, Zhou M, Arttamangkul S, Haugland RP (2000) Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. J Biochem Biophys Methods 42:137–151. https://doi.org/10.1016/s0165-022x(00)00048-8
Serra-Vinardell J, Sandler MB, Pak E, Zheng W, Dutra A, Introne W, Gahl WA, Christine Malicdan M (2020) Generation and characterization of four Chediak-Higashi Syndrome (CHS) induced pluripotent stem cell (iPSC) lines. Stem Cell Res. https://doi.org/10.1016/j.scr.2020.101883
Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W et al (2013) Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798. https://doi.org/10.1016/j.neuron.2013.05.029
Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, Rad L, Fitzgerald TW, Craig NL, Bradley A (2011) Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 39:e148. https://doi.org/10.1093/nar/gkr764
Park MA, Jung HS, Slukvin I (2018) Genetic engineering of human pluripotent stem cells using PiggyBac transposon system. Curr Protoc Stem Cell Biol. 47:e63. https://doi.org/10.1002/cpsc.63
Woodard LE, Wilson MH (2015) PiggyBac-Ing models and new therapeutic strategies. Trends Biotechnol 33:525–533. https://doi.org/10.1016/j.tibtech.2015.06.009
Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA (2008) Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci 28:6926–6937. https://doi.org/10.1523/jneurosci.0800-08.2008
Min Y, Xu W, Liu D, Shen H, Xu Y, Zhang S, Zhang L, Wang H (2013) Earle’s balanced salts solution and rapamycin differentially regulate the Bacillus Calmette-Guerin-induced maturation of human dendritic cells. Acta Biochim Biophys Sin (Shanghai) 45:162–169. https://doi.org/10.1093/abbs/gms117
Palmisano I, Della Chiara G, D’Ambrosio RL, Huichalaf C, Brambilla P, Corbetta S, Riba M, Piccirillo R, Valente S, Casari G, Mai A et al (2012) Amino acid starvation induces reactivation of silenced transgenes and latent HIV-1 provirus via down-regulation of histone deacetylase 4 (HDAC4). Proc Natl Acad Sci U S A 109:E2284–E2293. https://doi.org/10.1073/pnas.1202174109
Munson MJ, Allen GF, Toth R, Campbell DG, Lucocq JM, Ganley IG (2015) mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J 34:2272–2290. https://doi.org/10.15252/embj.201590992
Lattao R, Rangone H, Llamazares S, Glover DM (2021) Mauve/LYST limits fusion of lysosome-related organelles and promotes centrosomal recruitment of microtubule nucleating proteins. Dev Cell 56:1000-1013.e6. https://doi.org/10.1016/j.devcel.2021.02.019
Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY (2016) Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc 11:456–475. https://doi.org/10.1038/nprot.2016.018
Du W, Su QP, Chen Y, Zhu Y, Jiang D, Rong Y, Zhang S, Zhang Y, Ren H, Zhang C, Wang X et al (2016) Kinesin 1 drives autolysosome tubulation. Dev Cell 37:326–336. https://doi.org/10.1016/j.devcel.2016.04.014
Rong Y, McPhee CK, Deng S, Huang L, Chen L, Liu M, Tracy K, Baehrecke EH, Yu L, Lenardo MJ (2011) Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci 108:7826–7831. https://doi.org/10.1073/pnas.1013800108
Chang J, Lee S, Blackstone C (2014) Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J Clin Investig 124:5249–5262. https://doi.org/10.1172/jci77598
Rong Y, Liu M, Ma L, Du W, Zhang H, Tian Y, Cao Z, Li Y, Ren H, Zhang C, Li L et al (2012) Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol 14:924–934. https://doi.org/10.1038/ncb2557
Dai A, Yu L, Wang H-W (2019) WHAMM initiates autolysosome tubulation by promoting actin polymerization on autolysosomes. Nat Commun 10:3699. https://doi.org/10.1038/s41467-019-11694-9
McGrath MJ, Eramo MJ, Gurung R, Sriratana A, Gehrig SM, Lynch GS, Lourdes SR, Koentgen F, Feeney SJ, Lazarou M, McLean CA et al (2021) Defective lysosome reformation during autophagy causes skeletal muscle disease. J Clin Investig. https://doi.org/10.1172/jci135124
Schulze RJ, Weller SG, Schroeder B, Krueger EW, Chi S, Casey CA, McNiven MA (2013) Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol 203:315–326. https://doi.org/10.1083/jcb.201306140
Maday S, Holzbaur EL (2016) Compartment-specific regulation of autophagy in primary neurons. J Neurosci 36:5933–5945. https://doi.org/10.1523/jneurosci.4401-15.2016
Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15:1101–1111. https://doi.org/10.1091/mbc.e03-09-0704
Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147:728–741. https://doi.org/10.1016/j.cell.2011.10.026
Varga RE, Khundadze M, Damme M, Nietzsche S, Hoffmann B, Stauber T, Koch N, Hennings JC, Franzka P, Huebner AK, Kessels MM et al (2015) In vivo evidence for lysosome depletion and impaired autophagic clearance in hereditary spastic paraplegia type SPG11. PLoS Genet 11:e1005454. https://doi.org/10.1371/journal.pgen.1005454
Boutry M, Branchu J, Lustremant C, Pujol C, Pernelle J, Matusiak R, Seyer A, Poirel M, Chu-Van E, Pierga A, Dobrenis K et al (2018) Inhibition of lysosome membrane recycling causes accumulation of gangliosides that contribute to neurodegeneration. Cell Rep 23:3813–3826. https://doi.org/10.1016/j.celrep.2018.05.098
Khundadze M, Ribaudo F, Hussain A, Stahlberg H, Brocke-Ahmadinejad N, Franzka P, Varga R-E, Zarkovic M, Pungsrinont T, Kokal M, Ganley IG et al (2021) Mouse models for hereditary spastic paraplegia uncover a role of PI4K2A in autophagic lysosome reformation. Autophagy 17:3690–3706. https://doi.org/10.1080/15548627.2021.1891848
Dehay B, Bové J, Rodríguez-Muela N, Perier C, Recasens A, Boya P, Vila M (2010) Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci 30:12535–12544. https://doi.org/10.1523/jneurosci.1920-10.2010
Magalhaes J, Gegg ME, Migdalska-Richards A, Doherty MK, Whitfield PD, Schapira AHV (2016) Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: relevance to Parkinson disease. Hum Mol Genet 25:3432–3445. https://doi.org/10.1093/hmg/ddw185
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J-i, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884. https://doi.org/10.1038/nature04723
Marks MS, Heijnen HF, Raposo G (2013) Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol 25:495–505. https://doi.org/10.1016/j.ceb.2013.04.008
Ripoll L, Heiligenstein X, Hurbain I, Domingues L, Figon F, Petersen KJ, Dennis MK, Houdusse A, Marks MS, Raposo G, Delevoye C (2018) Myosin VI and branched actin filaments mediate membrane constriction and fission of melanosomal tubule carriers. J Cell Biol 217:2709–2726. https://doi.org/10.1083/jcb.201709055
Acknowledgements
The authors acknowledge National Human Genome Research Institute cytogenetics core for the iPSCs karyotyping; Stewart W. Humble (National Institute of Neurological Disorders and Stroke), Michael S. Fernandopulle (National Institute of Neurological Disorders and Stroke) and Jorge Gomez-Deza (Eunice Kennedy Shriver National Institute of Child Health and Human Development) for the technical advice. The authors also thank the Microfabrication and Microfluidics Unit of the Biomedical Engineering and Physical Science Shared Resource, National Institute of Biomedical Imaging and Bioengineering, for the assistance in the fabrication of the PDMS microdevices and templates; and Clàudia Serra Vinardell for her assistance in preparing graphic illustrations.
Funding
This study is supported by the Intramural Research Programs of the National Human Genome Research Institute, National Institute of Child Health and Human Development, National Institute of Allergy and Infectious Diseases and National Heart, Lung, and Blood Institute of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
JS-V, MBS, PS designing and conducting experiments, data analysis and interpretation, manuscript preparation-first draft, review, and editing. RDP helped with the trafficking experiments and data analysis. JM-L, JAB and AC contributed on microscopy data analysis. KK performed the bulk and single-cell sorting. WJI provided the CHS patients resources. MEW provided the i3N iPSC line and plasmids. WAG funding acquisition, review, and editing. MCCM review and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Serra-Vinardell, J., Sandler, M.B., De Pace, R. et al. LYST deficiency impairs autophagic lysosome reformation in neurons and alters lysosome number and size. Cell. Mol. Life Sci. 80, 53 (2023). https://doi.org/10.1007/s00018-023-04695-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04695-x