Abstract
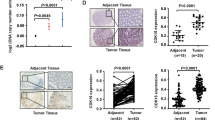
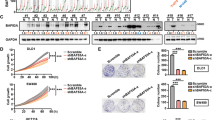
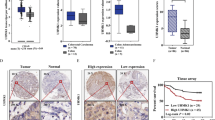
Upregulation of death-domain-associated protein (Daxx) is strongly associated with diverse cancer types. Among these, the clinicopathological significance and molecular mechanisms of Daxx overexpression in colorectal cancer (CRC) remain unknown. Here, we showed that Daxx expression was increased in both clinical CRC samples and CRC cell lines. Daxx knockdown significantly reduced proliferation activity in CRC cells and tumor growth in a xenograft model. Further studies revealed that Daxx expression could be attenuated by either treatment with the PIK3CA inhibitor PIK-75 or PIK3CA depletion in CRC cells. Conversely, expression of PIK3CA constitutively active mutants could increase Daxx expression. These data suggest that PIK3CA positively regulates Daxx expression. Consistently, the expression levels of PIK3CA and Daxx were positively correlated in sporadic CRC samples. Interestingly, Daxx knockdown or overexpression yielded decreased or increased levels of PIK3CA, respectively, in CRC cells. We further demonstrated that Daxx activates the promoter activity and expression of PIK3CA. Altogether, our results identify a mechanistic pathway of Daxx overexpression in CRC and suggest a reciprocal regulation between Daxx and PIK3CA for CRC cell growth.






Similar content being viewed by others
Availability of data and material
The data presented in this study are available from the corresponding author on reasonable request.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. CA Cancer J Clin 71:7–33
Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, Stivala F, Mccubrey JA, Libra M (2009) PIK3CA mutations in human solid tumors: role in sensitivity to various therapeutic approaches. Cell Cycle 8:1352–1358
Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA (2015) Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta 1855:104–121
Cancer Genome Atlas N (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330–337
Cathomas G (2014) PIK3CA in colorectal cancer. Front Oncol 4:35
Samuels Y, Waldman T (2010) Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol 347:21–41
Vanhaesebroeck B, Perry MWD, Brown JR, Andre F, Okkenhaug K (2021) PI3K inhibitors are finally coming of age. Nat Rev Drug Discov 20:741–769
Papadatos-Pastos D, Rabbie R, Ross P, Sarker D (2015) The role of the PI3K pathway in colorectal cancer. Crit Rev Oncol Hematol 94:18–30
Astanehe A, Finkbeiner MR, Hojabrpour P, To K, Fotovati A, Shadeo A, Stratford AL, Lam WL, Berquin IM, Duronio V, Dunn SE (2009) The transcriptional induction of PIK3CA in tumor cells is dependent on the oncoprotein Y-box binding protein-1. Oncogene 28:2406–2418
Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, Fernandez De Mattos S, Wymann MP, Brosens JJ, Schulze A, Lam EW (2008) The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol 28:5886–5898
Yang N, Huang J, Greshock J, Liang S, Barchetti A, Hasegawa K, Kim S, Giannakakis A, Li C, O’brien-Jenkins A, Katsaros D, Butzow R, Coukos G, Zhang L (2008) Transcriptional regulation of PIK3CA oncogene by NF-kappaB in ovarian cancer microenvironment. PLoS ONE 3:e1758
Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF 3rd, Maul GG (1999) PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–234
Xue Y, Gibbons R, Yan Z, Yang D, Mcdowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W (2003) The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A 100:10635–10640
Lin DY, Shih HM (2002) Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J Biol Chem 277:25446–25456
Shih HM, Chang CC, Kuo HY, Lin DY (2007) Daxx mediates SUMO-dependent transcriptional control and subnuclear compartmentalization. Biochem Soc Trans 35:1397–1400
Salomoni P, Khelifi AF (2006) Daxx: death or survival protein? Trends Cell Biol 16:97–104
Huang YS, Shih HM (2009) Daxx positively modulates beta-catenin/TCF4-mediated transcriptional potential. Biochem Biophys Res Commun 386:762–768
Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, Yang X (2006) Critical role for Daxx in regulating Mdm2. Nat Cell Biol 8:855–862
Kim EJ, Park JS, Um SJ (2003) Identification of Daxx interacting with p73, one of the p53 family, and its regulation of p53 activity by competitive interaction with PML. Nucleic Acids Res 31:5356–5367
Tsourlakis MC, Schoop M, Plass C, Huland H, Graefen M, Steuber T, Schlomm T, Simon R, Sauter G, Sirma H, Minner S (2013) Overexpression of the chromatin remodeler death-domain-associated protein in prostate cancer is an independent predictor of early prostate-specific antigen recurrence. Hum Pathol 44:1789–1796
Puto LA, Brognard J, Hunter T (2015) Transcriptional repressor DAXX promotes prostate cancer tumorigenicity via suppression of autophagy. J Biol Chem 290:15406–15420
Kwan PS, Lau CC, Chiu YT, Man C, Liu J, Tang KD, Wong YC, Ling MT (2013) Daxx regulates mitotic progression and prostate cancer predisposition. Carcinogenesis 34:750–759
Attieh Y, Geng QR, Dinardo CD, Zheng H, Jia Y, Fang ZH, Ganan-Gomez I, Yang H, Wei Y, Kantarjian H, Garcia-Manero G (2013) Low frequency of H3.3 mutations and upregulated DAXX expression in MDS. Blood 121:4009–4011
Zizzi A, Montironi MA, Mazzucchelli R, Scarpelli M, Lopez-Beltran A, Cheng L, Paone N, Castellini P, Montironi R (2013) Immunohistochemical analysis of chromatin remodeler DAXX in high grade urothelial carcinoma. Diagn Pathol 8:111
Yuen HF, Chan YP, Law S, Srivastava G, El-Tanani M, Mak TW, Chan KW (2008) DJ-1 could predict worse prognosis in esophageal squamous cell carcinoma. Cancer Epidemiol Biomark Prev 17:3593–3602
Lin GJ, Huang YS, Lin CK, Huang SH, Shih HM, Sytwu HK, Chen YW (2016) Daxx and TCF4 interaction links to oral squamous cell carcinoma growth by promoting cell cycle progression via induction of cyclin D1 expression. Clin Oral Investig 20:533–540
Chen SF, Kasajima A, Yazdani S, Chan MS, Wang L, He YY, Gao HW, Sasano H (2013) Clinicopathologic significance of immunostaining of alpha-thalassemia/mental retardation syndrome X-linked protein and death domain-associated protein in neuroendocrine tumors. Hum Pathol 44:2199–2203
Liu SB, Lin XP, Xu Y, Shen ZF, Pan WW (2018) DAXX promotes ovarian cancer ascites cell proliferation and migration by activating the ERK signaling pathway. J Ovarian Res 11:90
Tzeng SL, Cheng YW, Li CH, Lin YS, Hsu HC, Kang JJ (2006) Physiological and functional interactions between Tcf4 and Daxx in colon cancer cells. J Biol Chem 281:15405–15411
Huang YS, Chang CC, Lee SS, Jou YS, Shih HM (2016) Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget 7:43256–43266
Huang CJ, Chien CC, Yang SH, Chang CC, Sun HL, Cheng YC, Liu CC, Lin SC, Lin CM (2008) Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer. J Cell Mol Med 12:1936–1943
Chang CC, Naik MT, Huang YS, Jeng JC, Liao PH, Kuo HY, Ho CC, Hsieh YL, Lin CH, Huang NJ, Naik NM, Kung CC, Lin SY, Chen RH, Chang KS, Huang TH, Shih HM (2011) Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell 42:62–74
Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM (2006) Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24:341–354
Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE (2004) High frequency of mutations of the PIK3CA gene in human cancers. Science 304:554
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Ikegami D, Akiyama H, Suzuki A, Nakamura T, Nakano T, Yoshikawa H, Tsumaki N (2011) Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development 138:1507–1519
Huang YS, Chang CC, Huang TC, Hsieh YL, Shih HM (2012) Daxx interacts with and modulates the activity of CREB. Cell Cycle 11:99–108
Chang CC, Lin DY, Fang HI, Chen RH, Shih HM (2005) Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J Biol Chem 280:10164–10173
Khaiboullina SF, Morzunov SP, Boichuk SV, Palotas A, St Jeor S, Lombardi VC, Rizvanov AA (2013) Death-domain associated protein-6 (DAXX) mediated apoptosis in hantavirus infection is counter-balanced by activation of interferon-stimulated nuclear transcription factors. Virology 443:338–348
Pan WW, Zhou JJ, Liu XM, Xu Y, Guo LJ, Yu C, Shi QH, Fan HY (2013) Death domain-associated protein DAXX promotes ovarian cancer development and chemoresistance. J Biol Chem 288:13620–13630
Ko TY, Kim JI, Park ES, Mun JM, Park SD (2018) The clinical implications of death domain-associated protein (DAXX) expression. Korean J Thorac Cardiovasc Surg 51:187–194
Arafeh R, Samuels Y (2019) PIK3CA in cancer: the past 30 years. Semin Cancer Biol 59:36–49
Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I, Kurzrock R (2011) PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS ONE 6:e22769
Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R (2005) PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 65:2554–2559
Oda K, Okada J, Timmerman L, Rodriguez-Viciana P, Stokoe D, Shoji K, Taketani Y, Kuramoto H, Knight ZA, Shokat KM, Mccormick F (2008) PIK3CA cooperates with other phosphatidylinositol 3’-kinase pathway mutations to effect oncogenic transformation. Cancer Res 68:8127–8136
Vogt PK, Kang S, Elsliger MA, Gymnopoulos M (2007) Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem Sci 32:342–349
Park J, Lee JH, La M, Jang MJ, Chae GW, Kim SB, Tak H, Jung Y, Byun B, Ahn JK, Joe CO (2007) Inhibition of NF-kappaB acetylation and its transcriptional activity by Daxx. J Mol Biol 368:388–397
Acknowledgements
We thank Dr. Chi-Jung Huang and Mrs. Hsiao-Ting Tseng for CCD18-co cells, Dr. Noriyuki Tsumaki for PIK3CA reporter construct, Dr. Scott Lowe for MSCV Luciferase PGK-hygro plasmid, Dr. Feng Zhang for pX330 construct and Dr. Bert Vogelstein for PIK3CA H1047R and E545K plasmids, respectively. We are grateful to Dr. Shun-Yuan Jiang, Dr. Po-Hsun Tu, Dr. Wei-Chih Yang, and Mr. Pei-Hsiang Hou for their excellent technical support.
Funding
This work was supported by grants from the Ministry of Science and Technology (MOST 110-2327-B-001-001 and 108-2320-B-001-035-MY3), Taiwan.
Author information
Authors and Affiliations
Contributions
Conceptualization: YSH, CCW, and HMS; performing experiments: YSH, CCC, and HYK; data analysis: CCW and SFH; drafting and editing YSH and HMS.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared that they have no conflict of interests.
Ethical approval
The questionnaire and methodology for this study were approved by the Human Research Ethics committee of the Tri-Service General Hospital (Ethics approval number: 098-05-292). All animal handling was approved by the Ethical Committee on Animal Experiments, Academia Sinica, Taiwan (IACUC-approved ptotocol#2009-018).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
The participant has consented to the submission of these research findings to the journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, YS., Wu, CC., Chang, CC. et al. Reciprocal regulation of Daxx and PIK3CA promotes colorectal cancer cell growth. Cell. Mol. Life Sci. 79, 367 (2022). https://doi.org/10.1007/s00018-022-04399-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04399-8